Thanks Andras, I owe you so much.
Hi,
Sorry for these 2 new questions:
- How to save fiducial (F.fcsv) in python?
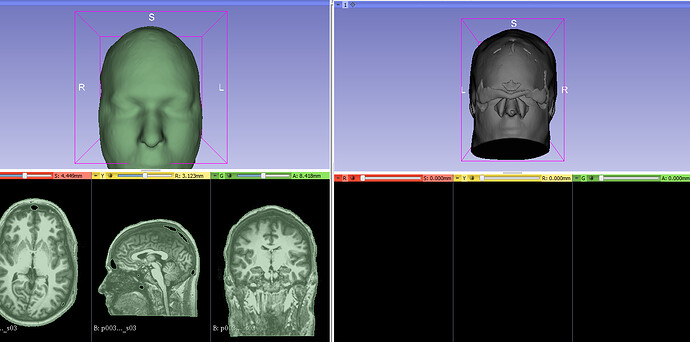
- With the wonderful script of @lassoan, I obtain great segmentation of my data (cf. my picture, left part), but I don’t know why there is some mistake during the saving of the stl (cf. my picture, rigth part). This mistake just occur with my data. Does someone have any idea what may cause this?
Thanks in advance.
- You can use this function from slicer.util https://github.com/Slicer/Slicer/blob/master/Base/Python/slicer/util.py#L423-L439
- There are holes in your segmentation. You’ll need to adjust the threshold parameters (likely reducing MinimumThreshold would help)
What mistake you are referring to?
Thanks @cpinter.
My data.stl was not like the segmentation mesh, they are all deformated (On my figure, the mesh and the data.stl are in the same point of view).
@lassoan I think he’s referring to the artifacts in the grey model that are caused by the holes.
@Frederic In your screenshot it can be seen that the segmentation is not solid (see the holes I was talking about), and those show up inside the head, looking quite strange. Once you fill the holes by adjusting the threshold parameters (or just filling them manually first with a brush to test my theory), the mesh on the right should look fine.
@cpinter I dont think that the cause is the holes, the segmentation of Andras script’s have holes too and the data.stl was fine.
My script is the same that Andras, I have just change:
effect.setParameter(“MinimumThreshold”,“100”)
effect.setParameter(“MaximumThreshold”,“2000”)
And one of my file was here (link).
Could you try with this file please?
The difference between the green and gray is that the green has full opacity, and the gray is semi-transparent, and full opacity hides the holes, semi-transparency lets them show. If you change the segmentation’s opacity to 50% or so, or change the model’s opacity to 100%, then they will look the same.
Thanks @cpinter , but It’s not that. Below, on the left my data (opacity: 100%, the holes are always here), on the right, the data of the script of Andras (opacity: 70%, with holes too).
I see. The problem is the surface normals. If you change Visible Slides to “Back-facing” in the Models module, then it will show up the same way

You can also fix the normals themselves in the Surface Toolbox module like this:

Great, you are right @cpinter ! really thanks!
And how to access to is module in python (to perform that you say following the script of Andras) please?
Surface Toolbox is a Python module, so you can copy-paste the few lines that updates the normal.
Yes. Unfortunately the module is poorly designed to be used more easily form the outside (for reference the “state” class is not exposed, and the UI widgets are not members).
Replace the last section of Andras’ code with this:
# Fix normals
surfaceMesh = segmentationNode.GetClosedSurfaceRepresentation(addedSegmentID)
normals = vtk.vtkPolyDataNormals()
normals.SetAutoOrientNormals(True)
normals.ConsistencyOn()
normals.SetInputData(surfaceMesh)
normals.Update()
surfaceMesh = normals.GetOutput()
# Write to STL file
writer = vtk.vtkSTLWriter()
writer.SetInputData(surfaceMesh)
writer.SetFileName("d:/something.stl")
writer.Update()Thanks to both of you once again. 
Hello,
Thanks for thoses informations. Do you know how to get the model properties like volume, surface, etc. With a python script ?
Is there a documentation for the python scripts?
Thank you very much!
You can use the Segment Statistics module to do that. See for example
Hi Andras,
I gone through the example segmention. I dont understand how to select the seed values? is it random selection? can it be automated as well?
tumorSeed.SetCenter(-6, 30, 28)
backgroundSeedPositions = [[0,65,32], [1, -14, 30], [0, 28, -7], [0,30,64], [31, 33, 27], [-42, 30, 27]]
You usually define seeds manually, using paint or draw effect. For fully automatic registration, you can define seeds on an atlas image; then you can transfer these seeds segments to a new image using registration (e. g., General registration (BRAINS) or General registration (Elastix) modules).
A post was split to a new topic: How to write STL from dicom slices