Is it useful to LekSell frame?
we do use:
i don´t know if the geometry (of fiducials) are the same.
the sides of each square are 140 mm.-
if not, you can modify the soft to get the proper calculations.
thank you ! I see, it is different, leksell’s each square is 120mm。
You can download and locally modify the extension to match your frame. Ideally, you would add a selector to allow choosing the frame.
you can change the stereotatic definition in script:
Stereotaxia/Recursos/Maquina_Russell_Brown.py
there is a class:
class Marco_Micromar():
“”“Clase para encapsular datos del marco MICROMAR TM-03B”“”
which have the 9 points that reference the specific frame .
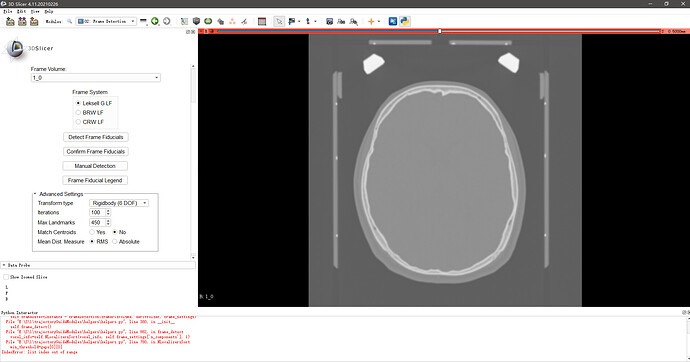
i have some sript error on use trajectoryGuideModules
when i press [Load Data] button ,nothing happen
Can you show me how you change the parameters?
i just change the parameter from 140 to 120

To correct some confusion… @jay1987 is uing my module trajectoryGuide and not Stereotaxia. @jay1987 to fix your error you need to first have your dataset in BIDS format: https://bids.neuroimaging.io/.
trajectoryGuide supports Leksell, BRW, and CRW frame systems.
thanks,i try to use slicer to positioning Leksell headframe , i use Stereotaxia and trajectoryGuide module , both encountered different problems.
in my process to use trajectoryGuide module
1.git clone trajectoryGuideModules from github site to local
2.follow the guide of site https://trajectoryguide.greydongilmore.com/installation.html to install module
3.use slicer to load dcm files and save it to nii.gz format

4.use Data Import module to load the nii.gz folder
5.change to Frame Detection module ,and select Leksell G LF to Detect Frame Fiducials,after seconds computing ,i got these errors
the detail information bellow
@Greydon_Gilmore
@jay1987 do you mind sharing the frame CT volume?
thanks from replying , i don’t known how to upload .rar data , it seems only support png,mp4 data uploading,would you mind leave your email for me ?
You can upload data to dropbox, OneDrive, Google drive, etc. and post the download link here.
thanks lasso,i made a dropbox link here
Dear jay1987:
Leksell and Micromar frames are different. So you can´t use the graphical issues (Stereotaxia module),
but the light version (StereotaxiaLite) can support the Leksell with the following changes :
Also: I´ve never worked with a Leksell. I don´t know if the zero in Z is the bottom of the fiducials (I suppose it is) and I don´t know where is the zero in Y ( I assumed its in the middle of the ruler)
fiducials is a square of 120 mm and they are separated 190

if you take an send me a CT-scan of a free Lexsell frame, with the fiducials visibles, I could make a model of it, take the meassures and implemet a version of Stereotaxia with it.
Dear JBeninca:
thanks for replying so much
i don’t have a leksell frame data now , only have a CT-SCAN with leksell head frame sequence here
Yes, i´ve downloaded it and delete the cranium and make a model of the frame. i think i could incorporate as an option in the module.
I must check some details about the frame: have you used it?
thanks
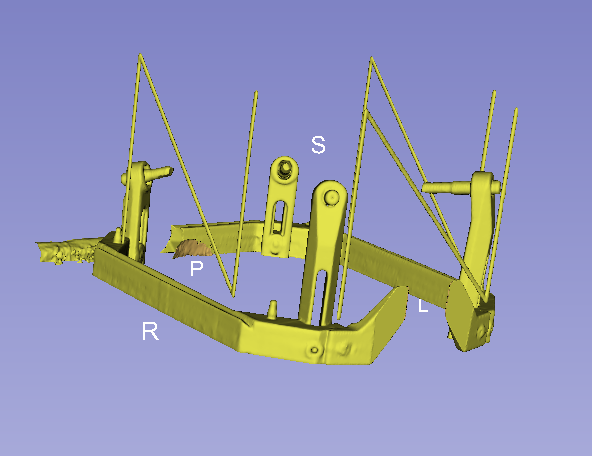
Leksell model from your CT scan
good job Beninca
Yes, my jobs is to location the head position by leksell head frame , and use it as a component for two hospital
can you integrate the leksell module in your Stereotaxia? it means much for us.