I have been working on 4D volume sequences (.nrrd format) on 3D slicer. I use segmentation editor to crop some parts where after I crop some parts I wanted to save it as another new volume. But after saving this I couldn’t open it as a 4D volume. It shows as a 3D volume without any 4D supports.
Segment editor can be used to create and edit segments. How did you use it to crop something? Do you mean you masked out certain region of an image volume?
Would you like to mask out a certain region in all frames of a volume sequence (4D volume) and save the result as a volume sequence?
Yes, I masked out certain region of volume sequence.I might be wrong. What I need to do eliminate some area from my 4D volume and save the results as 4D volume (volume sequence )
You need to create a new 4D volume sequence and go through your 4D volume and apply cropping to each frame:
Create a new 4D volume:
- In Segment Editor module, apply masking to the current frame of your volume (it’ll create a volume such as “CTP-cardio masked”)
- Go to “Sequence Browser” module, in Synchronized nodes section, click “+” icon => this creates a new sequence (while “Create new sequence” is selected on the left) and a new row appears in the table below
- In that new row, set the “Proxy node” to the masked volume (e.g., “CTP-cardio masked”) to associate the 3D volume with the 4D sequence, and check “Save changes” checkbox to automatically save modifications (when you mask the volume, the result will be saved in the sequence)
Apply masking to each frame:
- Go to Segment Editor module
- Click on Mask volume effect
- Click Apply
- Go to next frame by hitting Ctrl + Shift + Right-arrow key
- Click Apply
- … repeat the last 2 steps until you reach the end of your sequence
I know that the last two steps may be tedious (especially if you have hundreds of frames), but it can be automated using Python scripting. Let me know if you need this and I can describe how to do that.
Hi!
This problem is still relevant and I wonder if any progress on this topic has been carried out.
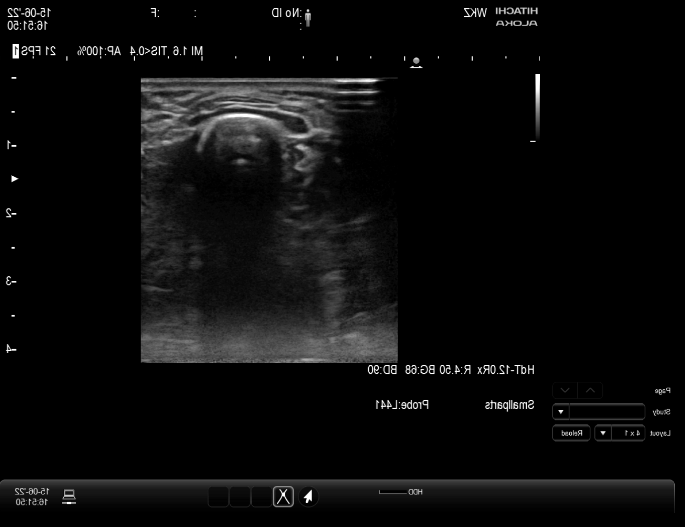
I have a live US stream to Slicer via the Plus Server and openIGTlink. However, my stream contains the whole US interface including all not useful aspects as visible in the picture below. The machine learning module is now performing the algorithm on this window instead of the US images alone. Is there any possibility to mask or crop the input signal in a live manner before getting a sequence or giving it to the live Unet algorithm?
Thanks!
You can crop 4D volume sequences using Crop volume sequence module. Let us know if you have any questions about how to use it.
Thanks for your answer!
Is it also possible to crop the input signal in the PuDeviceSet_Server_MmfVideoCapture.xml instead by just adding a few lines of code? That would be perfect as I am only interested in the
Yes, you can clip the image to the relevant region by setting ClipRectangleOrigin and ClipRectangleSize parameters in the Plus configuration file - see specification here.
Thanks for your answer. You really helped me out!
It worked with just adding a line of code ![]()
A little remark for the specification you sent me.
For two-dimensional input data the third component must be ommitted instead of may be ommitted in my case.
Thanks for confirming. I’ve updated the language around “may be” (changed to “may need to be”, as I’m not sure if it “must be” omitted for every devices).