why some dicom data are not cropped in 3d slicer? Once after setting the ROI and click apply button, its not cropping the selected region. Please help to resolve this issue.
Please explain in detail what you are trying to achieve, which 3D Slicer version you use, and which extension you use. Why is “using Python” in your title and not in your question?
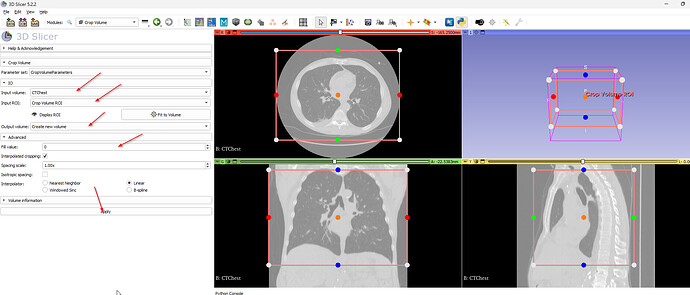
If you want to crop a volume in 3D Slicer 5.2.2 you will want to use the “Crop Volume” model. Create a new ROI. Set the parameters. Press “Apply”.
We are trying to crop the dicom to segment specific regions of brain. We can’t do the ROI based cropping for some of the datasets. We are able to set ROI and set parameters. But when we click the apply button it is not cropping the ROI region. We tried this both manually and with python scripting. But we couldn’t make it. Please help me.
GPT-4:
Here are a few potential solutions:
-
Check for software updates: Check if you have the latest stable version. If not, you may need to update or try a nightly build which might have the issue fixed.
-
Verify that the region of interest (ROI) is correctly defined: Make sure the ROI you’re trying to crop is correctly placed and defined in your data. If the ROI is outside the image boundaries or not properly aligned with the data, it could cause issues.
-
Investigate data issues: The problem could be with the data itself. It might be worth checking if there are any unusual features of the datasets where cropping isn’t working. For example, they could have different orientations, voxel sizes, or other metadata compared to the datasets where cropping works.
-
ROI not visible: It might be that your ROI is being cropped, but it’s not immediately visible in the view you’re looking at. Try adjusting the view settings or zooming out to see if the cropped ROI appears.
-
Reinstall 3D Slicer: If all else fails, you might need to reinstall 3D Slicer to make sure there are no issues with the installation.
In case you’re looking for a sample Python script to crop volumes based on ROI, here’s a basic example (untested)
# Get the current active volume node
inputVolume = slicer.mrmlScene.GetNodeByID('vtkMRMLScalarVolumeNode1')
# Create a new ROI node
roiNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLAnnotationROINode")
# Set the ROI to encompass the whole input volume
roiNode.Initialize(slicer.vtkMRMLAnnotationROINode.PlaceWidget, inputVolume)
# Now let's crop the volume
cropVolumeNode = slicer.modules.cropvolume.logic()
# Create a new output volume node
outputVolume = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLScalarVolumeNode")
# Set parameters
cropVolumeNode.SetInputVolume(inputVolume.GetID())
cropVolumeNode.SetROINode(roiNode.GetID())
cropVolumeNode.SetOutputVolumeNode(outputVolume.GetID())
# Apply cropping
cropVolumeNode.Apply()
Please replace 'vtkMRMLScalarVolumeNode1' with the ID of your current active volume node. Also, this script assumes that you want to crop the whole volume. You will need to adjust the ROI position and size if you want to crop a specific area. Please ensure you have the correct volume and ROI IDs.
Thank you for the previous questions and clarifying how to use Python script to crop volume. We manually created ROI regions and are trying to crop them using Python script. We assigned inputROI using getNode(“nameOfROIBox”) and used the code above to create a new outputVolume node. The problem we are having is with the inputVolume. How do we set inputVolume? When we tried using the code above, we had trouble knowing how to access the ID of our current active volume node.
Additionally, running these three lines of codes results in AttributeErrors saying the following…
Set parameters
cropVolumeNode.SetInputVolume(inputVolume.GetID())
cropVolumeNode.SetROINode(roiNode.GetID())
cropVolumeNode.SetOutputVolumeNode(outputVolume.GetID())
- ‘vtkSlicerCropVolumeModuleLogicPython.vtkSlicerCrop’ object has no attribute ‘SetROINode’
- ‘vtkSlicerCropVolumeModuleLogicPython.vtkSlicerCrop’ object has no attribute ‘SetInputVolume’
- ‘vtkSlicerCropVolumeModuleLogicPython.vtkSlicerCrop’ object has no attribute ‘SetOutputVolumeNode’
Thank you for helping us with this!
That would be because the ChatGPT-generated code supplied above is not correct. This slightly modified code from the Slicer script repository should get you started:
ct = getNode('myImageName')
roi = getNode('myROIName')
cropVolumeParameters = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLCropVolumeParametersNode")
cropVolumeParameters.SetInputVolumeNodeID(ct.GetID())
cropVolumeParameters.SetROINodeID(roi.GetID())
slicer.modules.cropvolume.logic().Apply(cropVolumeParameters)
croppedCT = cropVolumeParameters.GetOutputVolumeNode()
You may find a function here to crop volumes using Python.
To get any node in Slicer, use a qMRMLNodeComboBox as a GUI widget. You will have to go through the API to know how to use it.
If you just want to draw a ROI and crop a volume, you may consider implementing this project. It’s just copy and paste.
If you want to go deeper and do things yourself, you’ll have to go through scripting basics and get familiar with Slicer’s internals.
Good luck.
Thank you for providing reference to the script. The modified script was a tremendous help! The code worked. Now that we have the cropped volume from running the script, is there a way to write another line of code that automatically renames the obtained cropped volume for us?
This should work:
croppedCT.SetName('myNewName')
Hi again. We are integrating our code into Jupyter notebook, since the Python Console in Slicer is inconvenient to use directly. The code we previously had works in Slicer but now we have to adapt it to run in Jupyter notebook, and we encountered an error in recognizing the CT scan files we are working with. When we run the following lines of code, the second line is able to perform the getNode function on the given ROI. However, the first line gives us the following error message: MRMLNodeNotFoundException: could not find nodes in the scene by name or id ‘substack_name’
ct = slicer.util.getNode(“substack_name”)
roi = slicer.util.getNode(“ROI_name”)
What is the function to be able to get the substack in Jupyter notebook?