Hello,
I just began using slicer 3D, it is quite powerfull tool!
I am not using it for medical application, but in material science.
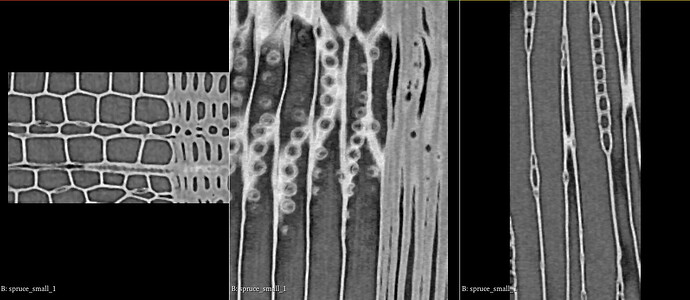
I have 3D scans from wood, that I would like to segment.
when the data has been imported it looks like this:
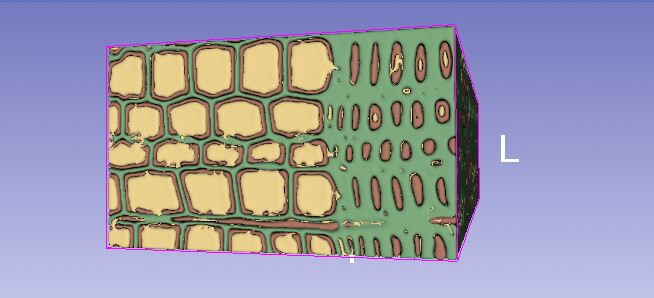
the data is for a small hexahedral section (cubish like) of the complete scan. using the threshold i was able to differentiate the three main volumes:
nevertheless, i have some issues with the results:
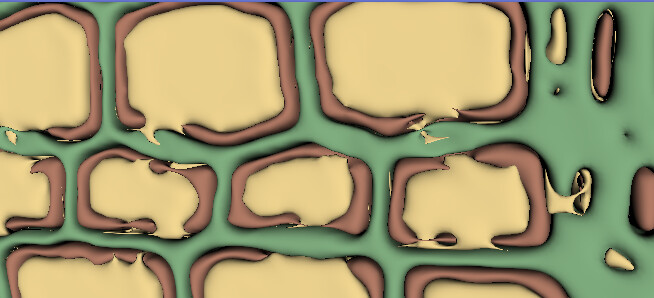
- in reality the 3 different regions, the black/red (frist picture/3D image) envelopes the grey/yellow one, and lastly the white/green comes. this images are the result from using threshold, but it can be seen that it is far from perfect:
but i dont know if this is possible to improve while using the threshold filter, I also tried painting and grow from seeds, option but did not gave good results (actually the best results i got where from threshold). any recommendations that someone could give me in this regards?
- I would like to have the different regions that the common boundaries are shared, i mean, in the surfaces that the regions are touching each other that the surface is the same, is this possible?
here is the data if anyone wants to play with.