Yes, I have. Not sure why it is not working
I think I figured it out–disabled and then enabled Slice Intersections seemed to do the trick. Many thanks again!
I have written a custom python script to do this (saved as a .py file, which I wrote in Notepad) but have not had luck running it with 3D Slicer. I added the path for the directory it is in to Edit–>Application Settings–>Modules and reset Slicer, but I don’t see it under the Examples menu.
See explanation in Slicer Python programming tutorial: https://www.slicer.org/wiki/Documentation/Nightly/Training#Slicer4_Programming_Tutorial
There is also a brand new option of running Slicer Python scripts from Jupyter notebooks.
I have followed the steps in the tutorial exactly, but I get an error when I try to add the module to the extension.

Check if the error log contains more details. Maybe module class name does not match exactly the .py file name.
Hello again, I have an additional question about this feature. Right now, I am attempting to calculate the total brain volume of a rat (excluding the cerebellum) from an MRI scan, using segmentation. I do this by creating a new segment, and painting the brain volume on each coronal slice (shown as lines against the oval-shaped axial cut below). I use the Segment Statistics module to view the total volume calculation (including all the slices) in mm^3.

My question is, how does Slicer calculate the total volume? If it starts from the top of the image and works downwards, I would imagine that the image below is a good representation of what is part of the volume calculation, with each of the lines representing the top edge of a given slice. Each rectangle is the width of the slice thickness, and I have left slight gaps to represent the gaps between the slices, which are not included in the volume calculation.

On the other hand, it could start from the bottom and work up, as shown below:
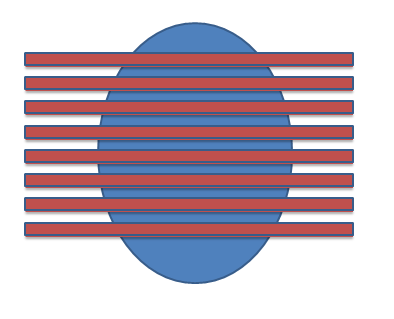
It is important for me to know how the volume is being calculated, as I need to know whether the slices include the cerebellum (at the bottom of the brain).
Just look at the segmentation in the other orthogonal views and see what is included in the segmentation. By default the segmentations will be binary labelmaps that fill the voxels of the master volume and there wouldn’t be any gap.
Thank you. One point of confusion still: as per the DICOM header, my slice thicknesses are 1mm, and the spacing between slices is 2mm. This means that there is a gap between slices of 1mm. Does the volume calculation done by the statistics module include this gap as part of the calculation? I had asked this question before and was assured that only the slice thickness (not the gap) was part of the volume calculation, but the 3D rendering of the segment appears to fill in the gap as well.
Thank you. One point of confusion still: as per the DICOM header, my slice thicknesses are 1mm, and the spacing between slices is 2mm. This means that there is a gap between slices of 1mm. Does the volume calculation done by the statistics module include this gap as part of the calculation? I had asked this question before and was assured that only the slice thickness (not the gap) was part of the volume calculation, but the 3D rendering of the segment appears to fill in the gap as well.
Hi @cphillips -
Perhaps it was misstated or unclear, but actually the the spacing between slices is the thing that Slicer uses for calculating volumes. The gap is not part of the calculation in the sense that Slicer is interpolating between slices and does not take into account the thickness and gap information.
To put it another way, Slicer constructs a volume which has a matrix IJKToRAS that maps from index space (IJK) to patient space (RAS). There’s no concept of a voxel being larger or smaller than the spacing. That is, if the thickness were larger than the slice spacing the slices would overlap. If it were smaller there would be a gap, but this is not directly represented.
Slice thickness indicates the quality of the imaging system - how well it can focus on a slice. The thickness value should not matter, unless extreme cases, e.g. if it is larger than the slice spacing or the size of the objects that you want to quantify.