I currently want to use 3D slicer to annotate 2D images. I have tried to convert the labeled results to binary labelmap and save them in dicom format. In this saving method, I can use python to read the labels correctly in the order of labels. However, there is a problem of overlapping labels, and the labels covered by subsequent labels are incompletely read. Then I tried to save the annotation result in nrrd format, and then read it with python, but the data format obtained by reading nrrd in this way is inconsistent, sometimes the size is (3wh1), sometimes it is (4w* h*1), and I did not find a definite correspondence between the pixel value and the corresponding label. How can I save the label file so that I can read out the labels of each category in turn, and there is no problem of incomplete labeling caused by overlapping labels.
Hi - to better help you, please try your workflow with the latest preview version and report the version number here along with the exact steps you took using public data so that anyone can reproduce. This sounds like a very solvable issue.
Thank you very much for your reply.
The version of the 3D Slicer I use is 4.11.20210226, and the code used to read the nrrd file is
data, _ = nrrd.read(path)
print(data.shape)
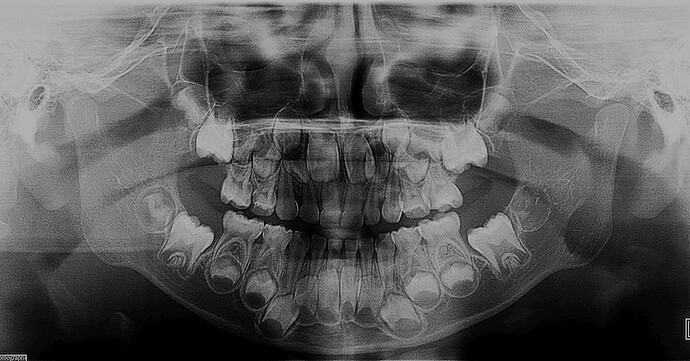
I also uploaded the original image and the annotated result. When I marked all the teeth on the original image, the shape obtained by reading the nrrd file using python was (4, 2440, 1280, 1), and when I only marked two teeth, the shape is (2, 2440, 1280, 1). And I can’t know the correspondence between the value on the read data and the label. When I save the annotation result in dicom format, the area of pixel value 1 on the read data represents the area marked by label 1. This correspondence is very clear, but saving as dicom cannot solve the problem of label overlap.
Non-overlapping labels can be stored in a single labelmap, but overlapping labels mean than there can be multiple labels at a single location, so a single labelmap is no longer sufficient to describe the relationship between labels and pixels (or voxels in the 3D case). When there is overlap between labels, Slicer creates additional layers of labelmap such that overlapping labels are on different layers of the labelmap. In your two-teeth example, two layers are required because each tooth must be on a separate layer because they overlap. More overlapping segments will generally require more layers, although not necessarily one per segment because it will typically be possible to put non-overlapping sets of segments on each layer. I don’t know what algorithm Slicer uses to sort segments into layers, but the result is 4 layers for your multi-tooth segmentation.
If you open the .nrrd file in a text editor (such as Notepad++) you will see that the top of the file is a human readable header. There will be a large section with information about each segment. You can see there each segment’s name and which layer of the label map it is stored on.
Unfortunately, I don’t know anything about DICOM segmentations or how Slicer exports to there, so I can’t help you with your problem there, but hopefully this is helpful as background information for understanding what is going on.
Thank you very much for your explanation. I will use the text editor to check the nrrd file. Have you used 3D Slicer to label overlapping labels before? Is there another way to solve the label overlap problem?
We developed the slicerio Python package to make it very easy to get pixels and metadata of overlapping segments from a .seg.nrrd file. You can use this Python package anywhere, not just in Slicer’s Python environment, by simply pip-installing slicerio.
Do you need DICOM segmentations? If not, and you were just viewing that as a way of exporting segmentations outside of Slicer, then @lassoan’s solution should work well for you. If you need the segmentations in DICOM format, then it would be helpful to have more information about what your goal is and what exactly is going wrong on the way to that goal.
I just want to convert 3D Slicer’s annotations to images, not necessarily in DICOM format, @lassoan 's solution is very effective and has solved my problem. Thank you all for your help.