Operating system: Mac
Slicer version: Slicer 4.10.0
Expected behavior: 4D NIFTI images load with an expected orientation of [1 0 0][0 1 0] [0 0 1]. If I concatenate multiple 3D datasets together and load the resulting the file the image orientation is reported as [-1 0 0][0 -1 0] [0 0 1]. I have verified the orientation with nifti_tool and found that it reports the same orientation between the two datasets.
Actual behavior:
Slicer uses RAS coordinate system internally, so if your input IJK to LPS matrix was [1 0 0][0 1 0] [0 0 1] then in Slice the IJK to RAS matrix is expected to be [-1 0 0][0 -1 0] [0 0 1]. Therefore, what you see may not be a problem.
How did you load the 4D Nifti file?
What is the 4th dimension? Time?
There’s been a lot of discussion of this topic.
I took a shot at fixing the MultiVolume Importer a few years ago and found it impossible to pin down any real standard for 4D nifti, but made things work for the files on the nifti web site. Beyond that I tend to throw up my hands and consider any other files application-specific and not interoperable.
@Vincent_Magnotta if you can make the case that there is a valid definition of 4D nifti that will allow true interoperability then for sure we would want Slicer to read and write those correctly, ideally via ITK. Things may have changed, but nfortunately last time I looked there was no such definition. Even 3D nifti has some issues.
The example that I have is from a project that is using NIFTI for handling fNIRS data. The data I am working with would be more similar to DTI data than fMRI data. A very simple example is that I take a 3D NIFTI image and simply run the
3dTcat command from afni that will concatenate two images together into a 4D image file. If I take the same image and concatenate the image with itself I end up with a NIFTI image that has the same NIFTI sform and qform transforms in the header. However, if
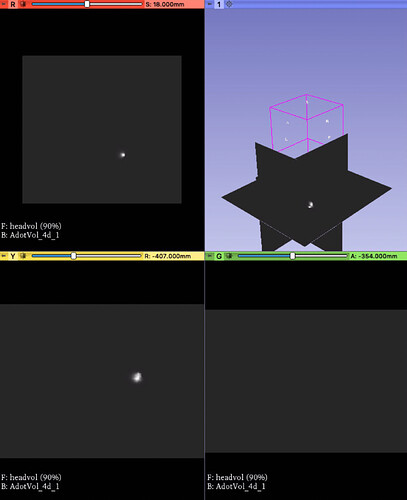
I load the 3D and 4D versions of the file into Slicer, the 3D image loads correctly and the 4D version that is loaded in a different orientation.
I can appreciate that NIFTI does not a great way to specify spatial and other dimensions of the data, but currently Slicer seems to at least interpret which dimensions are spatial relative to other dimensions. I would have expected that
the 3D and 4D versions would have had the same orientation and not be flipped relative to one another.
I agree, it’s frustrating for everyone.
Does it load correctly in ITK?
I did not try loading directly into an ITK image, but ITKSnap does load both images correctly and the results overlap as I would expected.
While investigating this orientation issue, you can apply an appropriate transformation matrix to fix the image after loading is completed; or you can use nrrd file format instead.
What is the 4th dimension in your data? Would you like to visualize it as a time sequence, a vector field, or just display a selected component? Would you like to process it? Using Segment Editor, or do some spatial registration, deformation, or process it using your own scripts using ITK/VTK/numpy,… ?
@Vincent_Magnotta can you post example data along with screen captures of what you think is the correct interpretation? Also can you provide links to the various apps you mention that generated the data along with the parameters you used. Sorry if this sounds super picky, but we’ve chased a lot of nifti orientation issues over the years and as I said earlier there is no clear standard for correctness.
Steve,
Totally understand the issues here. Happy to post some data. What is the preferred method to share? I am attaching a screenshot showing what I would expect the result to be and where the 4D image results when loaded. I am also attaching
the output of using nifti_tool to show the image headers.
For this I am using matlab and the associated NIFTI library to write the images files using the save_nii function. I then take a 3D image and concatenate it with itself to generate the 4D image, example command
3dTcat AdotVol_3d.nii AdotVol_3d.nii -prefix AdotVol_4d.nii
Output of nifti_tool
N-1 header file ‘headvol.nii’, num_fields = 43
all fields:
name
offset
nvals values
It could be google drive, onedrive, dropbox or whatever you think is most convenient.
And what method do you use to load them in Slicer? Which version of Slicer?
Another option, if you just use nifti to pass images between Matlab and Slicer to write 3D/4D nrrd files using nrrdwrite.m. NRRD files do not suffer from image orientation ambiguities and both 3D and 4D data sets are loaded consistently into Slicer.
What is the 4th dimension in your data? Would you like to visualize it as a time sequence, a vector field, or just display a selected component? Would you like to process it? Using Segment Editor, or do some spatial registration, deformation, or process it using your own scripts using ITK/VTK/numpy,… ? – The answers to these questions are important because you can represent 4D data various ways (vector volume, scalar volume with multiple components, multi-volume, or volume sequence) and different features are available for each.
Here is a link to the data.
https://www.dropbox.com/sh/g270f3mioz4u9vq/AAAxgfjdlfukGI1CVy65F4-pa?dl=0
I need these images to be in NIFTI format right now. We are working on an fNIRS pipeline that glues a lot of components together (HOMER/AtlasViewer, NeuroDot, AFNI, ANTS). I would really like to avoid conversion to another format. We have
worked hard to resolve all of the file format issues between these programs and NRRD just does not have the broad adoption to support this effort.
I fully agree, for the final solution you should be able to use any file format (and fix nifti specification and implementation wherever needed). I just recommended a workaround so you can proceed while it is being sorted out.
What is the 4th dimension in your data? Would you like to visualize it as a time sequence, a vector field, or just display a selected component? Would you like to process it? Using Segment Editor, or do some spatial registration, deformation, or process it using your own scripts using ITK/VTK/numpy,… ? – The answers to these questions are important because you can represent 4D data various ways (vector volume, scalar volume with multiple components, multi-volume, or volume sequence) and different features are available for each.