Hello,
I wanted to come back to this in case anyone was interested into craniosynostosis planning. Hopefully someone will find this useful!
My original goal was to re-create the workflow presented by García-Mato et al., but the Kitware Osteotomy Planner was not working as expected. Either way, I realized later that I personally like using the Transforms module more than using the local transformations/interactions the Kitware Osteotomy Planner uses.
Step-by-step:
-
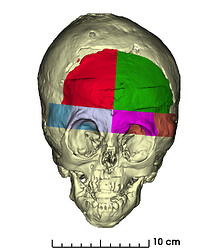
I used the Dynamic Modeler to recreate the Osteotomies as indicated by the Surgeon. I mostly used plane cuts for the osteotomies.
-
For relative translations of the bone segments I used the Transform module.
Advancement of the Orbital Segments, Superior View -
For local rotations, I had to create my own Axis of Rotation or Pivot Axis by creating a vertical Line on the midsagittal plane and coincident with the anterior edge of the advanced orbital segments. To perform the actual rotation, I then used this script from the Slicer doc.
Local Rotation of the Left Orbital Segments, Superior View
- I kept repeating steps (2-3) to recreate the motions and rotations indicated by the Surgeon. This required two additional pivot axes. We generated two different positions of the Bandeau, one more aggressive than the other.
- Experimental: Finally, we overlaid the Bandeau onto the soft-tissue segmentations to “see” the degree of improvements on the orbits. Would be nice to do something with displacement maps! (working on this now, will update later)
Challenges moving forth:
-
For every rotation I had to use the script from the doc. Is there a more elegant way of indicating a local rotation on the Transforms module? Is the solution to convert the script into a Python module/function that I just call every time? (instead of copying and pasting)
-
Is there a way of “linking” or “creating dependencies” on the transformations? My goal is to update the initial advancement (for instance) and allow for slicer to make all of the updates downstream, does that make sense?
-
Is there a way of using a Displacement Map generated from Model to Model Distance to then warp another model, say the soft tissue in step 5?
Thank you!