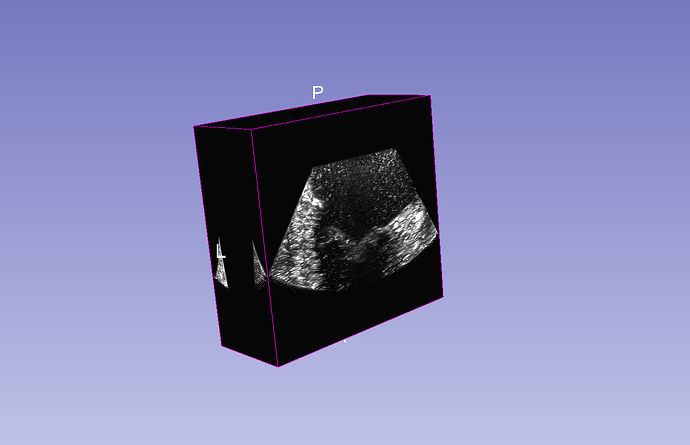
I had a look at the data set and tried all 3 methods.
Method C does not work because slices intersect each other and therefore cannot be interpolated using a grid transform.
Method B somewhat worked (I had to slightly improve the DICOM importer, the update will be available in tomorrow’s Slicer Preview Release). Since the the volume is quite sparse and the leaflet visibility is not always great, segmenting the leaflets like this can be quite challenging (you need to keep browsing the slices to get the necessary spatial and temporal context for interpreting the current slice).
Method A seems to be the most promising. Volume reconstruction module in SlicerIGSIO extension can manage to fill in the gaps between the slices and creates a full 3D volume, which can be visualized directly using volume rendering, segmented using the common segmentation tools, etc.
This is how the reconstructed 4D volume sequence looks like:
Farther from the mitral vale the gap between slices becomes too large, the holes cannot be filled anymore, but that could be addressed by changing the reconstruction parameters (and probably those areas are not interesting anyway).
You need to reconstruct the frames in groups, which requires reorganizing the sequence. Doing this manually for hundreds of frames would takes tens of minutes, so I wrote a script to automate this. You can load the MRI image series, define a ROI box to define where you want to reconstruct the volume, and run this script to get the result that is shown in the video above. It requires Slicer Preview Release downloaded tomorrow or later and installation of SlicerIGSIO and SlicerIGT extension.
# Set inputs
inputVolumeSequenceNode = slicer.util.getFirstNodeByClassByName('vtkMRMLSequenceNode', 'Sequence')
inputVolumeSequenceBrowserNode = slicer.modules.sequences.logic().GetFirstBrowserNodeForSequenceNode(inputVolumeSequenceNode)
inputVolumeSequenceBrowserNode = slicer.mrmlScene.GetFirstNodeByClass('vtkMRMLSequenceBrowserNode')
roiNode = slicer.util.getNode('R') # Draw an Annotation ROI box that defines the region of interest before running this line
numberOfTimePoints = 30
numberOfInstancess = 540
startInstanceNumbers = range(1,numberOfTimePoints+1)
# Helper function
def reconstructVolume(sequenceNode, roiNode):
# Create sequence browser node
sequenceBrowserNode = slicer.mrmlScene.AddNewNodeByClass('vtkMRMLSequenceBrowserNode', 'TempReconstructionVolumeBrowser')
sequenceBrowserNode.AddSynchronizedSequenceNode(sequenceNode)
slicer.modules.sequences.logic().UpdateAllProxyNodes() # ensure that proxy node is created
# Reconstruct
volumeReconstructionNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLVolumeReconstructionNode")
volumeReconstructionNode.SetAndObserveInputSequenceBrowserNode(sequenceBrowserNode)
proxyNode = sequenceBrowserNode.GetProxyNode(sequenceNode)
volumeReconstructionNode.SetAndObserveInputVolumeNode(proxyNode)
volumeReconstructionNode.SetAndObserveInputROINode(roiNode)
volumeReconstructionNode.SetFillHoles(True)
slicer.modules.volumereconstruction.logic().ReconstructVolumeFromSequence(volumeReconstructionNode)
reconstructedVolume = volumeReconstructionNode.GetOutputVolumeNode()
# Cleanup
slicer.mrmlScene.RemoveNode(volumeReconstructionNode)
slicer.mrmlScene.RemoveNode(sequenceBrowserNode)
slicer.mrmlScene.RemoveNode(proxyNode)
return reconstructedVolume
# This will store the reconstructed 4D volume
reconstructedVolumeSeqNode = slicer.mrmlScene.AddNewNodeByClass('vtkMRMLSequenceNode', 'ReconstructedVolumeSeq')
for startInstanceNumber in startInstanceNumbers:
print(f"Reconstructing start instance number {startInstanceNumber}")
slicer.app.processEvents()
# Create a temporary sequence that contains all instances belonging to the same time point
singleReconstructedVolumeSeqNode = slicer.mrmlScene.AddNewNodeByClass('vtkMRMLSequenceNode', 'TempReconstructedVolumeSeq')
for instanceNumber in range(startInstanceNumber, numberOfInstancess, numberOfTimePoints):
singleReconstructedVolumeSeqNode.SetDataNodeAtValue(
inputVolumeSequenceNode.GetDataNodeAtValue(str(instanceNumber)), str(instanceNumber),)
# Save reconstructed volume into a sequence
reconstructedVolume = reconstructVolume(singleReconstructedVolumeSeqNode, roiNode)
reconstructedVolumeSeqNode.SetDataNodeAtValue(reconstructedVolume, str(startInstanceNumber))
slicer.mrmlScene.RemoveNode(reconstructedVolume)
slicer.mrmlScene.RemoveNode(singleReconstructedVolumeSeqNode)
# Create a sequence browser node for the reconstructed volume sequence
reconstructedVolumeBrowserNode = slicer.mrmlScene.AddNewNodeByClass('vtkMRMLSequenceBrowserNode', 'ReconstructedVolumeBrowser')
reconstructedVolumeBrowserNode.AddSynchronizedSequenceNode(reconstructedVolumeSeqNode)
slicer.modules.sequences.logic().UpdateAllProxyNodes() # ensure that proxy node is created
reconstructedVolumeProxyNode = reconstructedVolumeBrowserNode.GetProxyNode(reconstructedVolumeSeqNode)
slicer.util.setSliceViewerLayers(background=reconstructedVolumeProxyNode)
slicer.modules.sequences.showSequenceBrowser(reconstructedVolumeBrowserNode)