Operating system:windows 10
Slicer version:4.8.0
Expected behavior: DTI Process
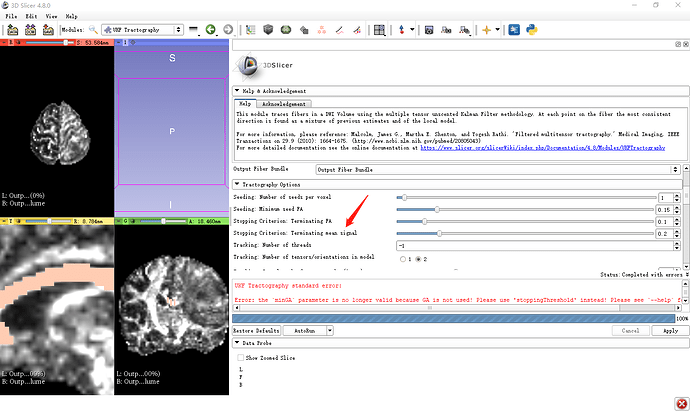
Actual behavior: Standard error
The current UKF Tractography is different from the tutorial(4.4)
Hi @Jin, thanks for the report. The minGA error has already been fixed in the 4.8.1 release, so please download that (or a more recent SlicerPreview build).
We will update the documentation.
Also, here’s an explanation of the change to UKF tractography. We have made this change (removing GA and adding the new parameter of normalized mean signal) to provide a measure that is suitable for both single-shell and multi-shell (multiple b-values) data.
I will try it, thanks very much!
Operating system: Ubuntu 18.04
Slicer version: 4.10
Is it possible to set waypoints or endpoints in the UKF tractography module?
Those mainly are used for probabilistic tractography. For UKF, the traditional processing is to seed tractography in a large area or in the whole brain, then select the tracts of interest. You can use regions of interest for selection, and these can correspond to the waypoints or endpoints you are interested in.
If you let us know your overall goal we may have more suggestions to help.
Hello @ljod
I was wondering if it is possible to perform seeding tractography with seed and endpoint. I am quite aware that streamline tractography needs only the seeding region. However, in my case, I want to add some endpoints as well to explore the tracts starting from one region and end at the specific region. I do have the seeding label and end label.
I am just not sure how to perform region-based stratigraphy. Is ROI selection in the diffusion module helpful? Can you guide me to how to perform this tractography?
Thank you.
I apologize for the late reply. We have tutorials about ROI selection here under Slicer Fiber Bundle Selection:
http://dmri.slicer.org/docs/