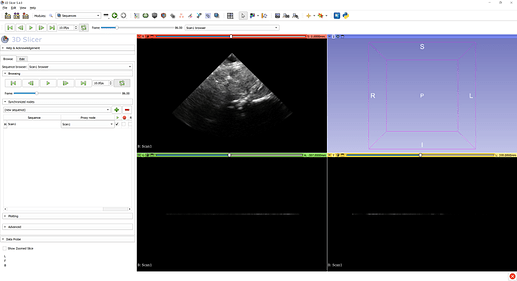
Hi, I’m trying to create a sequence file from a folder of ultrasound images in png format with poses stored in a csv file and then visualize in 3D.
I successfully created the .mhd header file containing the pose information and the poses can be visualized in Slicer. The file “Scan1.seq.mhd” looks like below:
ObjectType = Image
NDims = 3
AnatomicalOrientation = RAI
BinaryData = True
BinaryDataByteOrderMSB = False
CenterOfRotation = 0 0 0
CompressedData = False
ElementSpacing = 1 1 1
Offset = 0 0 0
TransformMatrix = 1 0 0 0 1 0 0 0 1
UltrasoundImageOrientation = MFA
UltrasoundImageType = BRIGHTNESS
Seq_Frame0000_ImageToReferenceTransform = 0.25880223 -0.96586309 0.01139754 99.35319738 -0.00294990 0.01100917 0.99993505 19.23409344 -0.96592583 -0.25881905 0.00000000 4.28620653 0.00000000 0.00000000 0.00000000 1.00000000
Seq_Frame0000_ImageToReferenceTransformStatus = OK
Seq_Frame0000_Timestamp = 0.000
Seq_Frame0000_ImageStatus = OK
Seq_Frame0001_ImageToReferenceTransform = 0.25868718 -0.96543371 0.03191713 99.30936271 -0.00826076 0.03082958 0.99949052 18.11578949 -0.96592583 -0.25881905 0.00000000 4.28620653 0.00000000 0.00000000 0.00000000 1.00000000
Seq_Frame0001_ImageToReferenceTransformStatus = OK
Seq_Frame0001_Timestamp = 0.300
Seq_Frame0001_ImageStatus = OK
Seq_Frame0002_ImageToReferenceTransform = 0.25844698 -0.96453726 0.05360077 99.21784580 -0.01387290 0.05177437 0.99856245 16.90844963 -0.96592583 -0.25881905 -0.00000000 4.28620653 0.00000000 0.00000000 0.00000000 1.00000000
Seq_Frame0002_ImageToReferenceTransformStatus = OK
Seq_Frame0002_Timestamp = 0.600
Seq_Frame0002_ImageStatus = OK
Seq_Frame0003_ImageToReferenceTransform = 0.25810672 -0.96326738 0.07414098 99.08820620 -0.01918910 0.07161468 0.99724777 15.78648773 -0.96592583 -0.25881905 -0.00000000 4.28620653 0.00000000 0.00000000 0.00000000 1.00000000
Seq_Frame0003_ImageToReferenceTransformStatus = OK
Seq_Frame0003_Timestamp = 0.900
Seq_Frame0003_ImageStatus = OK
…
ElementDataFile = Scan1.seq.nrrd
However, when I load this file into slicer as Sequence Metafile, it cannot pull the image data into the Slicer even though the Scan1.seq.nrrd file is under same directory as the .mhd metafile. I created the “Scan1.seq.nrrd” by opening the folder containing png images in slicer as volume, and save it as .nrrd file. I wonder if I should convert the png images into raw data file in a different way?
Also I wonder if I should set the ultrasound image spacing parameter in .mhd file? E.g., if my pixel spacing is 0.27mm, should I change the ElementSpacing = 0.27 0.27 0.27?
I appreciate any help!