import slicer
import qt
fname = qt.QFileDialog.getOpenFileName()
loadedVolumeNode = slicer.util.loadVolume(fname)
segfname = qt.QFileDialog.getOpenFileName()
segmentationNode = slicer.util.loadSegmentation(segfname)
labelmapNode = slicer.mrmlScene.AddNewNodeByClass('vtkMRMLLabelMapVolumeNode')
slicer.modules.segmentations.logic().ExportVisibleSegmentsToLabelmapNode(segmentationNode, labelmapNode, loadedVolumeNode)
parameters = {}
parameters["InputVolume"] = segmentationNode
parameters["Decimate"] = 0
parameters["JointSmoothing"] = False
parameters["SplitNormals"] = False
parameters["PointNormals"] = True
parameters["Pad"] = True
outputH = slicer.vtkMRMLModelHierarchyNode()
outputH.SetScene(slicer.mrmlScene)
outputH.SetName("3D Models")
slicer.mrmlScene.AddNode(outputH)
parameters["ModelSceneFile"] = outputH
parameters["Filter Type"] = 'Laplacian'
parameters["Smooth"] = 15
modelMaker = slicer.modules.modelmaker
slicer.cli.runSync(modelMaker, None, parameters)
ras2lps = vtk.vtkMatrix4x4()
ras2lps.SetElement(0,0,-1)
ras2lps.SetElement(1,1,-1)
ras2lpsTransform = vtk.vtkTransform()
ras2lpsTransform.SetMatrix(ras2lps)
# This is to try
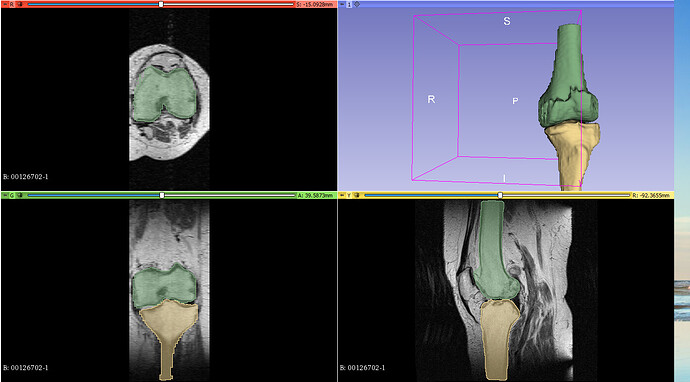
slicer.util.setSliceViewerLayers(background=loadedVolumeNode)
segmentationNode.setSliceViewerLayers(0)
When I set the segmentationNode.setSliceViewerLayers(0), the 3D model created using the above mentioned script also gets hidden. How can I decouple the 3D model and the segmentations. I would just like to visualize the MRI volumes and the 3D models.