how to load the image of single patient in dicom browser
Select folders that contain DICOM files
- Option A: Drag-and-drop the folder that contains DICOM files to the Slicer application window. Slicer displays a popup, asking what to do - click OK (“Load directory in DICOM database”). After import is completed, go to DICOM module.
- Option B: Go to DICOM module. Click “Import” button in the top-left corner of the DICOM browser. Select folder that contains DICOM files.
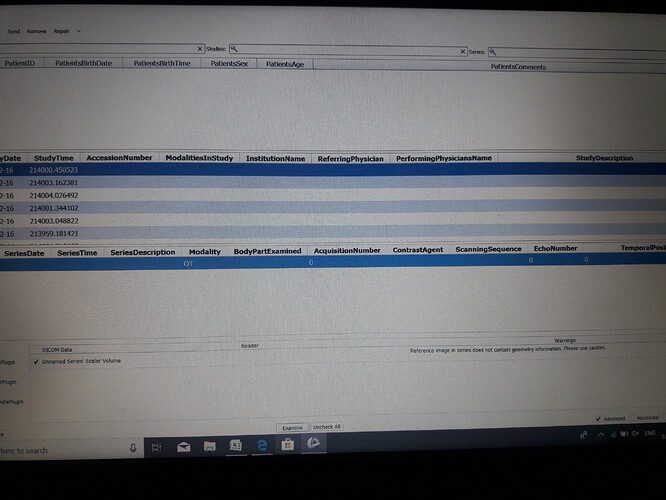
The selected study only contains “OT” = other data, something non-image. Use latest Slicer Preview Release and after the patient is imported, double-click on the patient in the DICOM browser to load all data that can be loaded for that patient.
i tried sir. but it didn’t work out
That error i get usually when there is spacing errors. In that case I load the data and inspect and i use dicom patcher to fix it. It has worked for me all the time. How ahead and look at the metadata and then load and see in the volume information whhere the problem is.
The patient name is not created for me sir…when i drag and drop the folder into the dicom browser…it shows 41 patients…like it sir…how to display the image in the screen of the 3d slicer software
These data sets have their modality set to “OT”, which means “other”. These might not images at all, or maybe they have an associated color image (a screenshot, a rendered image of a report, etc.). None of these are usable for visualization or analaysis.
Where do these files come from? What do you expect to have in them?
sir… our concept is that we are using the set of single patient mri brain images to convert into 3d reconstruction to identify the volume of the tumor.
The file that you showed the metadata content of appears to be a single 8-bit RGB screenshot without any geometry information (ConversionType=WSD means that “The equipment that captured the image from the screen, or placed the modified image into a DICOM SOP Instance”). Such images are not usable for any kind of analysis.
You need to export your data sets from the workstation as proper DICOM MR image series.
Thank you sir…i am final year student of kamaraj college of engineering and technology …i am doing final year project with the help of your software,which type of images can be used in 3d slicer software?please send sample mri brain image for our reference sir or send me any link to download images.
You have sample data that you can download within 3D Slicer. Just go to Sample Data module and you have sample data. MRI, CT etc…
Also when you start the 3D Slicer in the welcome screen you can see the link “Download Sample Data”
As for supported image/data formats can be found at
https://www.slicer.org/wiki/Documentation/Nightly/SlicerApplication/SupportedDataFormat
sir, how to calculate the volume of a 3d reconstructed mri brain image in 3d slicer software
First Segment the area you want to calculate. Use “segment editor” module for this,
You can find many tutorials online for this. Some common segmentation recipes can be found at
you can just apply the technique to your area of interest.
then you can use “segment statistics” module to to calculate volume and many other data.