Dear Dr. @fedorov,
Thank you so much for your kind information.
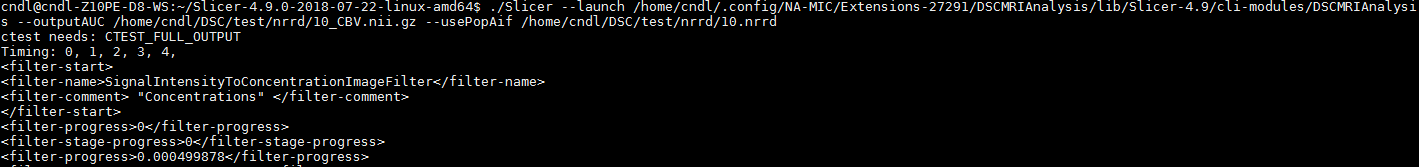
I managed to add some lines for the code, and the following result is acquired.
import os
import slicer
import MultiVolumeImporter
path1 = “/home/cndl/DSC/test/4d_nifti/”
path2 = “/home/cndl/DSC/test/nrrd/”
#os.mkdir(path2)
list = os.listdir(path1)
for filename in sorted(list):
importer = MultiVolumeImporter.MultiVolumeImporterWidget()
mvNode = slicer.mrmlScene.CreateNodeByClass(‘vtkMRMLMultiVolumeNode’)
slicer.mrmlScene.AddNode(mvNode)
importer.read4DNIfTI(mvNode, path1+filename)
mvNode.SetAttribute(‘MultiVolume.FrameIdentifyingDICOMTagName’,‘Time’)
mvNode.SetAttribute(‘MultiVolume.FrameIdentifyingDICOMTagUnits’,‘ms’)
mvNode.SetAttribute(‘MultiVolume.DICOM.FlipAngle’,‘1.00’)
mvNode.SetAttribute(‘MultiVolume.DICOM.EchoTime’,‘1.00’)
mvNode.SetAttribute(‘MultiVolume.DICOM.RepetitionTime’,‘1.00’)
fn_ext = os.path.splitext(filename)
true_fn_ext = os.path.splitext(fn_ext[0])
slicer.util.saveNode(mvNode, path2+true_fn_ext[0]+“.nrrd”)
exit()
(I’ve set all the additional information such as FlipAngle, EchoTime, and RepititionTime as ‘1.00’, which is default values of MultiVolumeImporter GUI.)
(Timing: values are seemed to be multiples of 0.00155, i.e. 0.00155 x 0, 1, 2, …, 95, which is the number of time points)
However, the “Timing:” seems a bit different from what I’ve got from the mpReviewPreprocessor.py.
(Timing:0,1,2,3,4,)
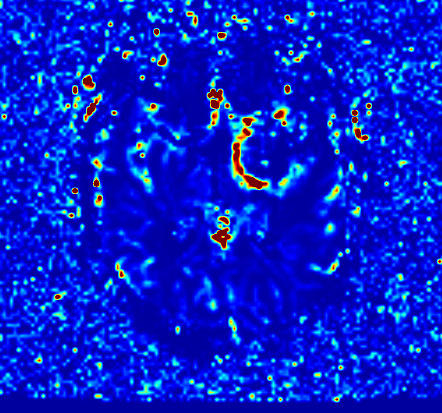
The resultant CBV map obtained from DSCMRIAnalysis is attached below.
-
with NRRD from read4DNIfTI
-
with NRRD from mpReviewPreprocessor.py
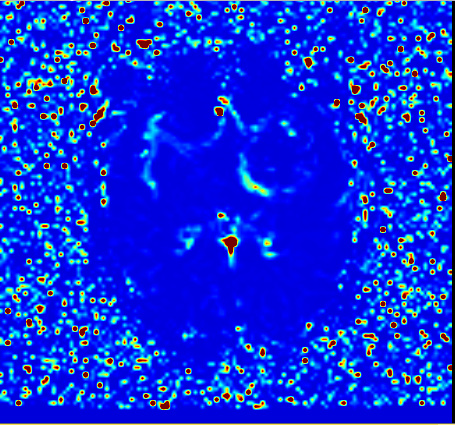
I think 1), and 2) images are a bit different.
Do you think it’s the problem of setting the FlipAngle, EchoTime, and RepititionTime as ‘1.00’?
However, I’ve changed them to 90,40,1900, which are true values from DICOM header information, and it didn’t make any difference.
Thank you in advance!
All the best,
Kyu Sung