Hello, we own both an E8 and BT.16 E10 And are planning to 3-D print to PLA for our customers. We have both types of volume formats,.VOL and .VOL Cartesian. We have read through these posts and have slicer set up with slicer heart. From what I can gather, we should be exporting.VOL Cartesian format, was that correct? We plan to then export as an .STL file or .OBJ file. What are the steps for the export of those file types? We would also be happy to send you any types of files from this equipment to help streamline the process
Yes.
You need to segment the volume using Segment editor module. See tutorials here. If you have any questions then post them as new topics.
Once you are done with segmentation, use “Segmentations…” button’s drop-down menu and choose “Export to files…”.
Thank you, that was helpful. I am referencing this video walkthrough but am running into some issues which are most likely user error, View your baby ultrasound and create 3D printable model using free software - YouTube
When I move from Volume Rendering to to Segment Editor, the segments I create are not shown in the 3d rendering box 1. Not sure what I am doing wrong.
This means of course that any edits I try to make will not take effect (such as scissors/magic cut)
You need to paint the mask (using threshold, paint, scissors, etc. effects) and use Mask Volume effect to apply the mask to the volume (blank out regions). Install SegmentEditorExtraEffects extension to get Mask volume effect in Segment Editor.
Really appreciate the help, that worked. How do you clear the 3D box when going from say Filtering/Volume Rendering/ ETC then to Segment Editor, I know I must be missing something simple?
You can show/hide the cropping ROI box in volume rendering module. You can show/hide any node in the Data module (subject hierarchy tab) by clicking on the eye icon.
If you have E10 you should be able to export STL directly from ultrasound.
I’ve actually had some success now after some deliberation to get this to work. I’m still having some issues getting the same resolution or quality that our E10 is getting. I am using the smoothing feature. If anyone else has any suggestions as far as improving the facial images for a later 3D print I’d love to hear it as I’m getting more familiar with this software.
I’m trying to print cardiac 3D volumes from TEE into slicer but i can’t load these files. I use GE Vivid E95 and E9. Any clue on how to load these files? Phillips cardio images load with no problems.
SlicerHeart can only read images exported in KretzFile format (or its DICOM-ized variant), which are mostly used by obstetrics and not cardiac systems. Last time we asked, GE would require us to sign a non-disclosure agreement to get specification of private DICOM fields that are required to read their TEE ultrasound volumes. However, such an agreement would prevent us from continuing our research and platform development work completely openly.
It would be great if GE changed their ultrasound systems to store image data in standard DICOM fields; or published how to retrieve image data from their private DICOM fields; or provided an export tool similar to Philips QLab (which creates DICOM files that store image data in standard fields). Make GE and your hospital administration aware of this issue and these potential solutions. Until GE does something about this, you may need to switch to Philips systems for research studies that require image data access.
I asked GE engineers about this issue and they don’t have an answer yet. How can I put the images of Philips platform in scale? The stl files became too small.
Philips systems store some essential information in private DICOM fields, therefore you need to follow these instructions to load them correctly: https://github.com/SlicerHeart/SlicerHeart/blob/master/README.md#philips
DICOMatic software claims to transform GE dicom into conventional dicom… Do you know if this archive can be loaded into slicer?
lassoan,
Thank you so much for your code. This really helps a lot.
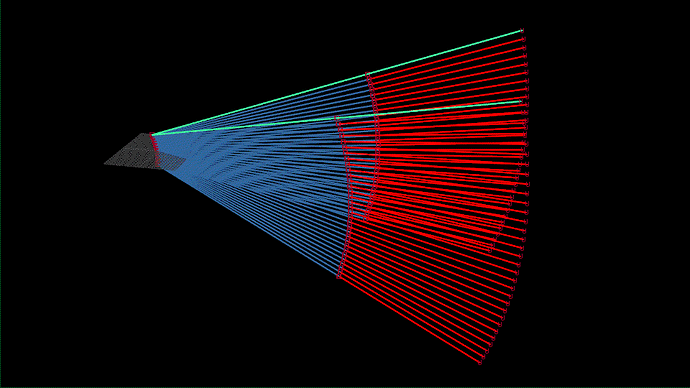
I tried visualizing the beam animation referencing your code. Here is the result.
This might be a question about the transducer’s mechanism rather than the code, but does the beam from 3D echo move like the above instead of like the following?
Or like the following?
I tried measuring some distances in the 3D space and of course the first one seems correct, but I didn’t know that the beam intersects during 3D echo. I looked for some technical document about the beam but I couldn’t find any materials…
Nice visualization!
Beamforming is partly mechanical (acoustic lens on the transducer) partly electronic (by adjustment of signal time delays), which allows a lot of design freedom. It is not necessary to use a single focal point (what would be the advantage? simpler reconstruction computations?). Instead, focusing is optimized for maximizing signal-to-noise ratio, penetration, geometrical accuracy, etc.
Note that these ultrasound files store a single spherical or cylindrical volume. In reality, the transducer probably generates many more beams, optimized for various depths and smartly integrates them. So, you won’t get too much insight into the inner workings of a transducer by inspecting these volumes.
Thank you so much for your reply.
If you do not mind, I would like to know how you formularized the most important part of your vtkSlicerKretzFileReaderLogic.cxx, from the line 280 to 282,
x = r * sin(theta)
y = -r * cos(theta) * sin(phi) + bModeRadius * sin(phi)
z = r * cos(theta) * cos(phi) + bModeRadius*(1-cos(phi))
espacially the latter part (bModeRadius * something at y, and z).
I could easily check that x and y was correct by measuring some length via the GE ultrasound apparatus, but I have no idea how you guessed the bModeRadius part.
The three animation gifs I made have been made by changing the bModeRadius part a little bit.
(The first gif is using your original formula.)
And I wonder if (C200, 0004), and (C200, 0005) values are unnecessary for reconstruct the volumes in 3D space.
Thank you.
I inspected many images and tried to find a logical explanation of how various inputs contributed to the reconstructed output. It was quite fun, like solving a riddle.
Hi András, would you be able to share the transferred files used for this model?
See download link in this post above.