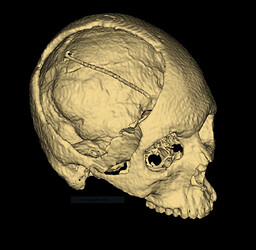
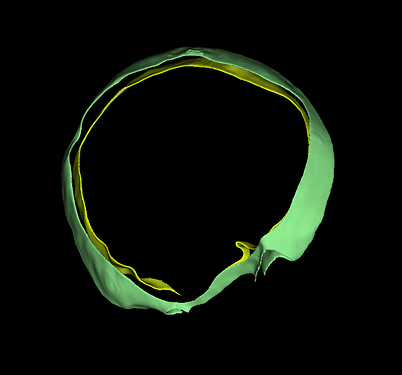
In case anyone is still using medial surfaces for thickness measurement and calculation, I have been working on a consolidated workflow to measure the thickness of cranioplasty implants;
Starting with a model of the implant (or any input surface mesh for that matter), the ThicknessMapping function (below) will complete all of the steps discussed previously, with the addition of some display settings:
For this input model;
The following medial thickness map is generated;
The program generates a blank voxel volume around the model, instead of using the original volume from the reconstruction. This reduces the number of voxels to be processed to calculate the medial surface. For standard 3.0 mm Head CTs I was originally waiting for about 10-13 minutes for the BinaryThinningImageFilter to complete. For this model shown here, the entire program takes about 1-2 mins 
Here is the program, written as a python function. Feel free to use and please give feedback!!!
def ThicknessMapping(self):
'''
Thickness Mapping
Generates and display the thickness of a model throughout its surface.
The thickness map is calculated using a combination of ITK Filters available to Slicer in python
References
[1] How to run a CLI module from Python (https://slicer.readthedocs.io/en/latest/developer_guide/python_faq.html?highlight=CLI#how-to-run-a-cli-module-from-python)
[2] Running an ITK filter in Python using SimpleITK (https://slicer.readthedocs.io/en/latest/developer_guide/script_repository.html#running-an-itk-filter-in-python-using-simpleitk)
[3] Create custom color map and display color legend (https://slicer.readthedocs.io/en/latest/developer_guide/script_repository.html#create-custom-color-map-and-display-color-legend)
'''
# Flags
verbose = True
# UI Inputs
thicknessMapInputModel = slicer.mrmlScene.GetNodeByID( self._parameterNode.GetNodeReferenceID("ThicknessMapInputModel") )
# Process
## Creating Function Directory
shNode = slicer.mrmlScene.GetSubjectHierarchyNode()
folder = shNode.CreateFolderItem(shNode.GetSceneItemID(), "Thickness Mapping")
## Get Model Centroid and Bounding Box
inputAssemblyBounds, assemblyBoundingBoxCentroid = self.logic.getBoundingBoxCentroid( thicknessMapInputModel.GetName(), True )
## Generate a Blank, Bounding Volume
masterVolumeNodeName = "{}_volume".format( thicknessMapInputModel.GetName() )
N = 2.0
imageSpacing = [1/N,1/N,1/N]
imageSize = [int(np.round(inputAssemblyBounds[1]-inputAssemblyBounds[0])*1.10*N),
int(np.round(inputAssemblyBounds[3]-inputAssemblyBounds[2])*1.10*N),
int(np.round(inputAssemblyBounds[5]-inputAssemblyBounds[4])*1.10*N)]
voxelType = vtk.VTK_UNSIGNED_CHAR
imageOrigin = [(assemblyBoundingBoxCentroid[0]-imageSize[0]/(2*N)),
(assemblyBoundingBoxCentroid[1]-imageSize[1]/(2*N)),
(assemblyBoundingBoxCentroid[2]-imageSize[2]/(2*N))]
imageDirections = [[1,0,0], [0,1,0], [0,0,1]]
fillVoxelValue = 0
imageData = vtk.vtkImageData()
imageData.SetDimensions(imageSize)
imageData.AllocateScalars(voxelType, 1)
imageData.GetPointData().GetScalars().Fill(fillVoxelValue)
masterVolumeNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLScalarVolumeNode", masterVolumeNodeName)
masterVolumeNode.SetOrigin(imageOrigin)
masterVolumeNode.SetSpacing(imageSpacing)
masterVolumeNode.SetIJKToRASDirections(imageDirections)
masterVolumeNode.SetAndObserveImageData(imageData)
masterVolumeNode.CreateDefaultDisplayNodes()
masterVolumeNode.CreateDefaultStorageNode()
shNode.SetItemParent( shNode.GetItemByDataNode(masterVolumeNode), folder )
# Status
if (verbose):
print( "Generated Blank, Bounding Volume..." )
## Generate a LabelMap using the Input Model and the Blank Bounding Volume
modelLabelMapParams = {}
modelLabelMapParams["InputVolume"] = masterVolumeNode
modelLabelMapParams["surface"] = thicknessMapInputModel
modelLabelMapNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLLabelMapVolumeNode","{}_labelmap".format( thicknessMapInputModel.GetName() ))
modelLabelMapParams["OutputVolume"] = modelLabelMapNode
modelToLabelMap = slicer.modules.modeltolabelmap
cliNode = slicer.cli.runSync(modelToLabelMap, None, modelLabelMapParams)
if cliNode.GetStatus() & cliNode.ErrorsMask:
errorText = cliNode.GetErrorText()
slicer.mrmlScene.RemoveNode(cliNode)
raise ValueError("CLI execution failed: " + errorText)
slicer.mrmlScene.RemoveNode(cliNode)
shNode.SetItemParent( shNode.GetItemByDataNode(modelLabelMapNode), folder )
shNode.SetItemParent( shNode.GetItemByDataNode(cliNode), folder )
# Status
if (verbose):
print( "Generated LabelMap from Model..." )
## Generating Medial Surface
inputImage = sitkUtils.PullVolumeFromSlicer( modelLabelMapNode )
filter = sitk.BinaryThinningImageFilter()
outputImage = filter.Execute( inputImage )
medialSurfaceVolumeNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLScalarVolumeNode", "{}_medial_surface".format( thicknessMapInputModel.GetName() ))
sitkUtils.PushVolumeToSlicer(outputImage, medialSurfaceVolumeNode)
shNode.SetItemParent( shNode.GetItemByDataNode(medialSurfaceVolumeNode), folder )
# Status
if (verbose):
print( "Generated Medial Surface from LabelMap..." )
## Generating Distance Map
inputImage = sitkUtils.PullVolumeFromSlicer( medialSurfaceVolumeNode )
filter = sitk.DanielssonDistanceMapImageFilter()
filter.UseImageSpacingOn()
filter.InputIsBinaryOn()
outputImage = filter.Execute( inputImage )
distanceMapVolumeNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLScalarVolumeNode", "{}_distance_map".format( thicknessMapInputModel.GetName() ))
sitkUtils.PushVolumeToSlicer(outputImage, distanceMapVolumeNode)
shNode.SetItemParent( shNode.GetItemByDataNode(distanceMapVolumeNode), folder )
# Status
if (verbose):
print( "Generated Distance Map from Medial Surface..." )
## Probe Volume with Model
probeVolumeWithModelParams = {}
probeVolumeWithModelParams["InputVolume"] = distanceMapVolumeNode
probeVolumeWithModelParams["InputModel"] = thicknessMapInputModel
thicknessMapNode = slicer.mrmlScene.AddNewNodeByClass("vtkMRMLModelNode","{}_thickness_map".format( thicknessMapInputModel.GetName() ))
probeVolumeWithModelParams["OutputModel"] = thicknessMapNode
probeVolumeWithModel = slicer.modules.probevolumewithmodel
cliNode = slicer.cli.runSync(probeVolumeWithModel, None, probeVolumeWithModelParams)
if cliNode.GetStatus() & cliNode.ErrorsMask:
errorText = cliNode.GetErrorText()
slicer.mrmlScene.RemoveNode(cliNode)
raise ValueError("CLI execution failed: " + errorText)
slicer.mrmlScene.RemoveNode(cliNode)
thicknessMapInputModel.GetDisplayNode().SetVisibility(False)
shNode.SetItemParent( shNode.GetItemByDataNode(thicknessMapNode), folder )
shNode.SetItemParent( shNode.GetItemByDataNode(cliNode), folder )
# Status
if (verbose):
print( "Generated Thickness Map from Distance Map and Input Model..." )
## Updating Display Settings, Colormap Labeling
# First, we set the medial surface volume on the background
slicer.util.setSliceViewerLayers( background = medialSurfaceVolumeNode, foreground = None, label = None )
# Here we ensure the thickness model is shwon intersecting all planes
thicknessMapNode.GetDisplayNode().SetSliceIntersectionVisibility(True)
thicknessMapNode.GetDisplayNode().SetSliceIntersectionThickness(3)
# And finally we plot the colormap;
# To generate the colormap, we need to extract the scalar range associated with the model
scalars = vtk_to_numpy( thicknessMapNode.GetPolyData().GetPointData().GetScalars() )
maxThickness = np.max( scalars )
minThickness = np.min( scalars )
avgThickness = np.mean( scalars )
colorTableTitle = "Medial Thickness (mm)"
labelFormat = "%4.1f mm"
colorTableRange = maxThickness
# Create color node
colorNode = slicer.mrmlScene.CreateNodeByClass("vtkMRMLProceduralColorNode")
colorNode.UnRegister(None) # to prevent memory leaks
colorNode.SetName(slicer.mrmlScene.GenerateUniqueName("MedialThicknessMap"))
colorNode.SetAttribute("Category", "MedialThicknessModule")
# The color node is a procedural color node, which is saved using a storage node.
# Hidden nodes are not saved if they use a storage node, therefore
# the color node must be visible.
colorNode.SetHideFromEditors(False)
slicer.mrmlScene.AddNode(colorNode)
# Specify colormap
colorMap = colorNode.GetColorTransferFunction()
colorMap.RemoveAllPoints()
colorMap.AddRGBPoint(0.0, 0.66, 0.0, 1.0)
colorMap.AddRGBPoint(0.5, 1.0, 0.0, 1.0)
colorMap.AddRGBPoint(1.0, 1.0, 0.33, 1.0)
colorMap.AddRGBPoint(1.5, 1.0, 1.0, 1.0)
colorMap.AddRGBPoint(2.0, 0.33, 1.0, 1.0)
colorMap.AddRGBPoint(2.0, 0.0, 1.0, 1.0)
colorMap.AddRGBPoint(maxThickness, 0.0, 0.66, 1.0)
# Display color legend
thicknessMapNode.GetDisplayNode().SetAndObserveColorNodeID(colorNode.GetID())
colorLegendDisplayNode = slicer.modules.colors.logic().AddDefaultColorLegendDisplayNode(thicknessMapNode)
colorLegendDisplayNode.SetTitleText(colorTableTitle)
colorLegendDisplayNode.SetLabelFormat(labelFormat)
# Status
if (verbose):
print( "Thickness Mapping Completed...!" )