We are looking for a histogram of the DVF.
The DVF should not have any negative values, but when I calculated the histogram from the DVF created by the transform module, it took negative values.
What do these negative values mean?
Also, I am converting a REG file created by MIMmaestro into a vector field by the transform module.
Will this procedure create the same creation in 3DSlicer as the one created in MIMmaestro?
I hope you can answer my question.
It is expected that the displacement vector field contains negative values. Only the displacement magnitude is always postitive.
You can compare the difference between two displacement fields by loading them as transforms, invert one of them and apply it on the other.
Thank you for your answer.
Is it correct to say that the DVF is transformed correctly in the transform module?
I am sorry for my lack of knowledge, but I would like to know about the negative value of DVF.
I didn’t have an image of DVF having a negative value because it is a vector shift amount.
I would like to know how DVF is defined in 3DSlicer and how negative values are generated.
I hope you can answer my question.
We believe so, yes.
Deformation vector fields are represented as “grid transforms” in Slicer, a special case of general transforms where there is an image-like set of regularly spaced vectors defining the deformation field. Each vector defines a local displacement in patient space, the components of which can be positive or negative to indicate the local displacement.
Thank you for your answer.
It was helpful because I was not sure if the method of converting the grid to DVF in the transform module was correct.
What do you mean by “the components of which can be positive or negative to indicate the local displacement.” I know that DVF takes 0 if nothing changes, but I don’t know what negative DVF means.
Please let me know if you can help me.
I hope you can answer my question.
The key point is what Andras mentioned above. The deformation vector field itself can have positive or negative components (three values: left/right, front/back, up/down) but the magnitude, or length of the vector or amount of movement, is a single value that is always positive or zero if there is no displacement.
Thank you for your answer.
I see that 3DSlicer determines the positive and negative values for left/right, front/back, and up/down.
If so, I think the voxel value displayed in the histogram will be the sum of the three positive and negative values, but what does the negative value mean when the three are combined?
I do not understand the idea of the negative value of the three elements combined.
fig:DVFhistogram

Slicer just provide all the vectors in a numpy array and you combine the values. So, the behavior is whatever you implement.
If you are interested in plotting the displacement magnitude then you can compute the norm of the vectors before computing the histogram. Since the processing of numpy arrays using numpy is unrelated to Slicer, you should be able to find solution on Stackoverflow or various generic Python tutorials and forums.
Thank you for your answer.
So you are saying that the result of summing the three axes of the numpy array, not just one axis of the histogram, is a positive or negative value.
Out of the blue, clinically speaking, does a negative value of DVF represent volume shrinkage and a positive value represent expansion?
If you know, please let me know.
No, the positive or negative elements of the displacement vector do not individually tell you about shrinking or expansion in a clinical sense.
For that you would use the determinant of the Jacobian matrix to give you a localized measurement. But that signal is probably very noisy in clinical images so people typically use overall volume change from a segmented image or a change in length of the maximum diameter.
Thank you for your answer.
So, are negative values of DVF in 3DSlicer just the result of the sum of the three axes and have no particular clinical significance?
I don’t think so, no. It’s not clear to me what you would use that kind of plot for.
I am trying to get a histogram of the DVF, which I think will help me understand how the segments were changed by the DIR (scaling up, down, translating).
I thought about the Jacobian, but I started with the DVF, which is the result of the DIR itself.
Then I thought that a negative value of DVF might represent a contraction of DIR.
Doesn’t I make much sense to check the DVF?
I am embarrassed to say this, but I would like to know if there is any mistake.
I ask again, does this mean that the calculation of the 3 axes to find the DVF value is out of 3DSlicer’s jurisdiction?
Average translation in a region of interest is easy to compute: it is simply the mean computed for each displacement component. “Scaling up/down” of a region is also easy to get, by using volume or bounding box metrics provided by Segment Statistics module.
There are extensions for more detailed, quantitative analysis, including computing Jacobian determinant and various statistics. See for example RegistrationQA extension.
You can also do any custom quantitative analysis in a few lines of Python code, using any Python packages, numpy, ITK, VTK, etc.
Thanks for the answer.
I used the segment statistics module to get the displacement field (DVF) statistics from Scalar volume statistics. Does this result show positive as expansion and negative as contraction?
The bounding box says “Oriented bounding box: the non-axis aligned bounding box that encompasses the segment”. Is this also suitable for interpreting DVF changes?
Let me add a question to my previous answer.
I believe that the DVF value of a histogram is represented by the sum of the squares of the three axes.
If negative values are generated during the calculation process, are the negatives meaningless?
If you know anything about this, please let me know.
Segment statistics only give meaningful information on scalar volumes, not on vector volumes.
Yes, by comparing the bounding box of the segments that you get with/without applying the displacement field transform.
I don’t understand this sentence. A histogram does not have a DVF value.
You can compute histogram for one component at a time. You can use “Vector to scalar volume” module to extract a single scalar component from a vector volume.
You can export a DVF magnitude image from a transform using Transforms module’s Convert section.
What is your overall goal? What is the clinical problem that you are trying to solve?
What does “I don’t understand this sentence. A histogram does not have a DVF value.” mean?
I am trying to get a histogram of the DVF by contour to see how it changed before and after the DIR.
So, I used the transform module to create a DVF and get the histogram for each contour of that VectorField.
I thought the DVF was the square root of the sum of squares of the three axes x,y,z, so I don’t understand why the histogram showed negative values.
I asked this because you wrote that “I believe that the DVF value of a histogram is…”, but a histogram does not have a DVF (displacement vector field) value. Each histogram value contains the number of values that are in the corresponding value range.
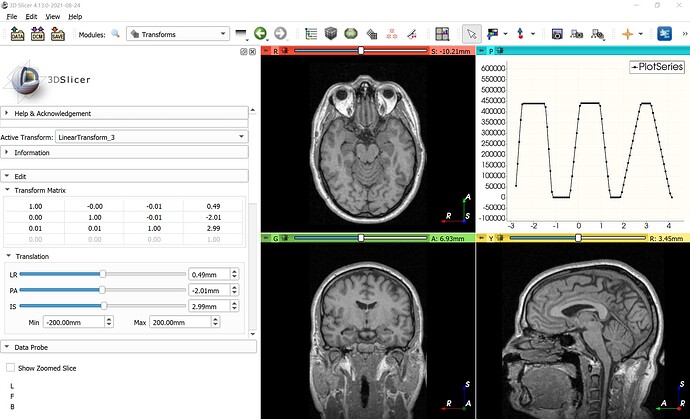
If you pass all the coordinate values to the numpy histogram function then it will simply pool together the values of all three vector components (R, A, S). Probably your histogram above have 3 peaks because displacement vector components are mostly around -2, -0.6, and 0.4.
To give you an example, if you create a transform with a translation around RAS axes [0.5, -2, 3] and add a few tenth of a degree of rotation (just to have some variance in the translation) then you get a histogram like this if you pool all the components together:
volumeNode = getNode('Displacement Field')
import numpy as np
histogram = np.histogram(arrayFromVolume(volumeNode), bins=100)
slicer.util.plot(histogram, xColumnIndex = 1)
Since statistics of all 3 vector components are pooled (thrown into one big list), you don’t know which peak correspond to which scalar component. So, you probably want to compute histogram for each component separately. You can then see the distribution of each vector component separately: R component’s peak is around 0.5, A component peaks around -2.0, S component around 3.0.
range = [-5.0, 5.0]
bins = 100
histogramR, histogramBins = np.histogram(arrayFromVolume(volumeNode)[:,:,:,0], bins, range)
histogramA = np.histogram(arrayFromVolume(volumeNode)[:,:,:,1], bins, range)[0]
histogramS = np.histogram(arrayFromVolume(volumeNode)[:,:,:,2], bins, range)[0]
slicer.util.plot(np.vstack([histogramBins[:-1], histogramR, histogramA, histogramS]).T, xColumnIndex = 0, columnNames=['N', 'R', 'A', 'S'], title='Histogram')
If you want to compute the histogram of the displacement magnitude then you will ignore vector directions and just compute statistics for the length of each vector. These numbers will be all positive. Since vectors are all about [0.5, -2.0, 3.0], the peak will be around 3.6:
magnitudes = np.linalg.norm(arrayFromVolume(volumeNode), axis=3)
histogram = np.histogram(magnitudes, bins=100)
slicer.util.plot(histogram, xColumnIndex = 1, columnNames=['magnitude', 'N'], title="displacement")
Note that all that I described above are just standard numpy functions and basic linear algebra. The only Slicer-specific functions are arrayFromVolume to get the displacement field vectors as a numpy array, and slicer.util.plot to display a plot.
Thanks for the answer.
I would like to evaluate each contour, where in the above code should I put the code you gave me before?
https://slicer.readthedocs.io/en/latest/developer_guide/script_repository.html#get-histogram-of-a-segmented-region
I tried everything, but it didn’t work.
I apologize for the inconvenience, but I would appreciate your advice.
I don’t know which code your are referring to and if you don’t provide the code that you tried and the error message you got then I cannot help figuring it what was wrong.
If you want to get displacement vectors in a specific segment then use the masked numpy array instead of the numpy array returned directly by arrayFromVolume.