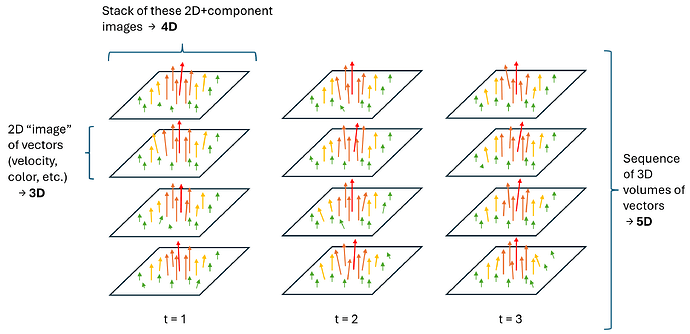
3D Slicer has expanded its capabilities to support reading and writing images, image sequences, and transform sequences with dimensionality up to 5D. This is a significant enhancement that enables users to work with larger and more complex imaging data, and do it more efficiently.
The following data types can now be stored in NRRD file format and loaded into Slicer for visualization and analysis:
- 3D
- Volumetric image (xyz)
- 2D image time sequence (xyt)
- 2D color image (cxy)
- 4D
- Color volumetric image (cxyz) - for example cryosection images (such as the visible human data set) or multichannel/multimodal images (such as B-mode + Color Doppler)
- Displacement fields (vxyz) - warping transforms
- Flow fields (vxyz) - such as 4D flow MRI
- Sequence of volumetric images (xyzt) - commonly used in cardiac echo, CT, MR or dynamic imaging

- Sequence of 2D color images (cxyt) - color video recording

- 5D
- Sequence of color volumetric images (cxyzt)

- Sequence of displacement fields (vxyzt) - sequence registration results

- Sequence of flow fields (vxyzt) - such as sequence of 4D flow MRI images

- Sequence of color volumetric images (cxyzt)
New use cases
The following use cases are enabled by these new developments.
Efficient storage of deformable image sequence registration results
Modules such as “Sequence Registration” have been available for motion tracking or motion compensation in 3D image sequences. However, after they computed a deformable transformation sequence, the only option to store it was a zip file containing several displacement field files. This was very inflexible and inefficient, as it was not possible to access a random item in the sequence and files were large.
In the current version of 3D Slicer, 4D displacement fields are stored by default as a single standard 5D NRRD file. The file can be saved as uncompressed raw data for fastest access; or compressed for maximum storage space efficiency.
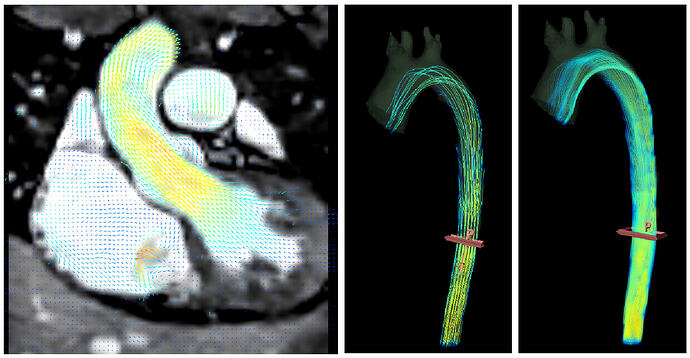
4D flow MRI
Some recent MRI systems allow real-time recording of 4D+t flow images, which store a 3D velocity vector at each voxel position, for tens of timepoints in the cardiac cycle. Previously, the only way to save such data sets was to save multiple 4D volumes in a zip file. These can now be saved as 5D vector field files.
Color video storage
3D Slicer has been supporting storage of color video recordings, but only at certain resolutions and only in 2D. The new infrastructure allows color images and image sequences to be stored efficiently in a single 5D image file.
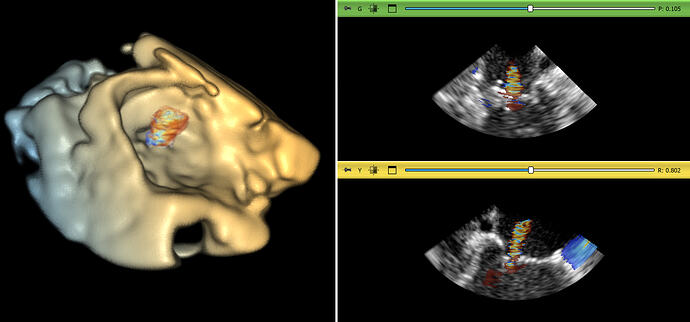
Multi-modality image storage
Cardiac ultrasound systems can acquire 3D B-mode ultrasound image time sequences, which were stored as 4D image volumes. Some systems can simultaneously acquire B-mode and Color Doppler (velocity) images. Since the resolution and field of view of these images are very similar, they can be robustly and efficiently displayed in 3D as a multi-component volumetric image. However, previously there was no way of storing a sequence of these fused multi-component volumetric images efficiently in one data file.
Usage examples
Using 3D Slicer
Slicer’s Python API makes it easy to load, manipulate, and save high-dimensional data.
Load any data type of any dimension (image, transform, image sequence, transform sequence):
node = slicer.util.loadNodeFromFile('path/to/file.nrrd')
Save any data type of any dimension (image, transform, image sequence, transform sequence):
slicer.util.saveNode(node, 'path/to/file.nrrd')
For sequences of images or transforms a common convention in Slicer is to use the composite file extension .seq.nrrd. This is not required by the data reader, but makes it easier for users to distinguish sequences from simple 3D files.
Using ITK Python
The new features are available in ITK Python 6.x.
Read a 5D displacement field vector file:
filename = "path/to/file.nrrd"
import itk
nrrd_io = itk.NrrdImageIO.New()
nrrd_io.SetAxesReorderToUseNonListRangeAxisAsPixel()
nrrd_io.SetFileName(filename)
nrrd_io.ReadImageInformation()
PixelType = itk.Vector[itk.F, nrrd_io.GetNumberOfComponents()]
dimension = nrrd_io.GetNumberOfDimensions()
ImageType = itk.Image[PixelType, dimension]
reader = itk.ImageFileReader[ImageType].New()
reader.SetFileName(filename)
reader.SetImageIO(nrrd_io)
reader.Update()
image = reader.GetOutput()
print(image.shape)
print(meta["NRRD_kinds[pixel]"])
Write the displacement field vector to file:
filename = "path/to/file.nrrd"
nrrd_io = itk.NrrdImageIO.New()
writer = itk.ImageFileWriter[ImageType].New()
writer.SetFileName(filename)
writer.SetImageIO(nrrd_io)
writer.SetInput(image)
writer.Update()
Using Python with pynrrd
Writing high-dimensional numpy arrays with correct metadata is fairly straightforward. Example for writing a time sequence of 3D volumes:
import numpy as np
import nrrd
img = np.zeros([201,201,201, 2])
header = {
'space': 'left-posterior-superior',
'kinds': ['domain', 'domain', 'domain', 'list'],
'labels': ["", "", "", "time"],
'units': ["", "", "", "s"],
'space origin': [-137.16099548, -36.80649948, -309.71899414],
'space directions': [[1.953125, 0., 0.], [0., 1.953125, 0.], [0., 0., 1.953125], [np.nan, np.nan, np.nan]],
'axis 3 index type': 'numeric',
'axis 3 index values': "1.2 1.4"
}
nrrd.write("c:/tmp/test.seq.nhdr", img, header)
The resulting image header:
NRRD0004
# Complete NRRD file format specification at:
# http://teem.sourceforge.net/nrrd/format.html
type: double
dimension: 4
space: left-posterior-superior
sizes: 201 201 201 2
space directions: (-1.9531249999999998,0,0) (0,-1.9531249999999998,0) (0,0,1.9531249999999998) none
kinds: domain domain domain list
labels: "" "" "" "time"
units: "" "" "" "s"
endian: little
encoding: gzip
space origin: (137.16099548000003,36.806499479999999,-309.71899414000001)
data file: test.seq.raw.gz
DataNodeClassName:=vtkMRMLScalarVolumeNode
axis 3 index type:=numeric
axis 3 index values:=1.2 1.4
Information for developers
We chose NRRD file format for storage of 5D information, because this format provides standard fields for specifying the meaning of each axis, it is a very simple file format, and it allows storing any number of custom fields for describing any details of the data that cannot be captured by standard fields.
To make it easy to read/write these higher-dimension files correctly in a wide variety of software, not just in 3D Slicer, we decided to implement this functionality in the ITK toolkit. This allows any software that uses ITK to read/write such files without any custom development. All Python developers can use this implementation, too, since ITK is available as a Python package. It is feasible to use the file format for any software that cannot use ITK, as writing of valid 5D files is fairly straightforward; and reading of specific variants can be implemented with not too much effort.
ITK C++ implementation
An ITK image can be an Image object that can store scalar, RGB, CovariantVector, etc. pixels in an arbitrarily high dimensional array. If the number of components of the pixel has to be dynamic then the ITK image has to be a VectorImage object that can store multi-component pixels in an arbitrarily high dimensional array.
NRRD file can contain domain (space, time) axes and range (list, RGB, covariant-vector, etc.) axes in an arbitrary order. Therefore, axes of the NRRD file may have to be reordered to be able to read the NRRD file into an ITK image. In general, NRRD domain axes are mapped to ITK image axes then all range axes are used as additional image dimensions. The only exception is that it makes sense to pick one range axis and use that as pixel component.
File Reading
Previously, a rigid, arbitrary logic was used for mapping higher-dimensional array to ITK data type. Now NRRD IO allows having more than one non-spatial axis and a few different strategies are avaialable to map them to ITK image types via the new AxesReorder property in NrrdImageIO.
The non-spatial axes of the NRRD file can be used in the ITK image as pixel components or as additional image dimensions.
The default strategy (UseAnyRangeAxisAsPixel) implements the same behavior as before (first non-spatial NRRD axis always becomes pixel component).
If the NRRD file contains a non-list type (e.g., RGB, point, covariant-vector) range axis then the first one is used as pixel component; the image is read into an Image object. If the NRRD file only contains list-type range axis then it is used as pixel component; the image is read into a VectorImage object. However, this resulted in more complicated application code (developers had to instantiate an Image or VectorImage depending on what kind of axes are in the NRRD file) and potential unnecessary reordering of axes (e.g., sequence of 3D images was always converted into a single multi-component 3D image).
The new UseNonListRangeAxisAsPixel option allows “list” dimension (e.g., time points) to always mapped to additional image dimension.
This new option simplifies application implementation, as time sequences are loaded the same way for scalar and vector volumes.
An additional UseScalarPixel option was introduced that is useful for cases when developers always want scalar pixel type. In this case, The image is always read into an ITK Image object, and all range (i.e., non-domain) axes of the NRRD file are added as additional domain axes.
Examples:
NRRD file with these axes kinds: list domain domain domain RGB-Color (or: domain domain domain list RGB-Color)
UseAnyRangeAxisAsPixel:Image<RGBPixel<double>,4>with axes: domain domain domain listUseNonListRangeAxisAsPixel:Image<RGBPixel<double>,4>with axes: domain domain domain listUseScalarPixel:Image<double,5>with axes: domain domain domain list RGB-Color
NRRD file with these axes kinds: list domain domain domain (or: domain domain domain list)
UseAnyRangeAxisAsPixel:VectorImage<double,3>with axes: domain domain domainUseNonListRangeAxisAsPixel:Image<double,4>with axes: domain domain domain listUseScalarPixel:Image<double,4>with axes: domain domain domain list
File writing
Previously the NRRD writer handled one component axis, and all the other dimensions were space.
Now it is possible to designate one of the axes as a list, by setting NRRD_kinds[i]=list metadata.
Example:
Previously, the file header for a 5D array was written as follows (4 domain axes):
dimension: 5
space dimension: 4
sizes: 4 320 320 1 5
space directions: none (1,0,0,0) (0,1,0,0) (0,0,1,0) (0,0,0,1)
kinds: RGBA-color domain domain domain domain
encoding: gzip
space origin: (0,0,0,1)
We can now write a file header like this (with 3 domain axes + 1 list axis):
dimension: 5
space: left-posterior-superior
sizes: 4 320 320 1 5
space directions: none (-1,0,0) (0,-1,0) (0,0,1) none
kinds: vector domain domain domain list
encoding: gzip
space origin: (0,0,0)
Axis units
NRRD IO did not support the “units” metadata for axes, now it is added both for reading and writing.
Per-axis metadata (kinds, labels, units) was only saved for spatial axes, now it is added for all axes.
For pixel components, pixel is used instead of axis index (NRRD_labels[pixel] and NRRD_units[pixel] fields).
Transforms
If only standard NRRD file fields are used, then displacement fields may not be distinguishable from other kind of vector fields.
To give a hint to applications that a NRRD file is intended to be used as a displacement field, the custom intent_code field is set to the value of 1006. This field and enumerated value comes from NIFTI file format specification. It simplifies applications that may read displacement fields from either NRRD or NIFTI formats.
Implementation in Slicer
The ITK classes described above are used in 3D Slicer in volume and transform storage nodes. Since 3D Slicer uses VTK pipelines, a new pair of classes, vtkITKImageSequenceReader and vtkITKImageSequenceWriter were implemented to wrap the ITK classes in a VTK interface. These classes are used in vtkMRMLVolumeSequenceStorageNode and vtkMRMLTransformSequenceStorageNode for image and transform storage.
Acknowledgments
This work has been funded in part by CHOP Frontier Program - Children’s Hospital of Philadelphia Frontier Programs conduct visionary research that translates to cutting-edge care; Additional Ventures 993932 - Computational Modelling of the Atrioventricular Valve Repair Single Ventricle Patients with Atrioventricular Canal, and NIH 1R01HL153166-01 - Computer Modeling of the Tricuspid Valve in Hypoplastic Left Heart Syndrome (PI: Matthew Jolley) driven by the SlicerHeart project.