when I try and import my .nrrd files as sequences (which they work in slicer 5.8.1) i get an error that slicer cannot read it. They will import as volume but then there is no way to change the colour map, i have tried the way you do in slicer 5.8.1. When i change the colour map it only change the legend bar. The only time the actual colours change is when i click on the presets, it always automatically imports as the rainbow scheme.
Thanks for reporting. Perhaps it’s related to these changes, @cpinter ?
@gchampa can you share a sample dataset that loads differently between the two versions?
It seems quite likely that it is related.
@gchampa Please include the entire message, because without that we don’t have any chance to make concrete suggestions.
Also, given that this is about reading a certain data file, if you can share the file, or any file that produces the same issue, then we’ll have everything to investigate and suggest a solution. Thank you!
yup ive added the nrrd file to onedrive: dose_distribution and the output when I try to upload as sequence is
- Error: Loading /home/gchampagne/Simulations/dose_distribution.nrrd - Error reading file.
- Error: Loading /home/gchampagne/Simulations/dose_distribution.nrrd - Failed to read node dose_distribution (vtkMRMLSequenceNode1) from filename=‘/home/gchampagne/Simulations/dose_distribution.nrrd’
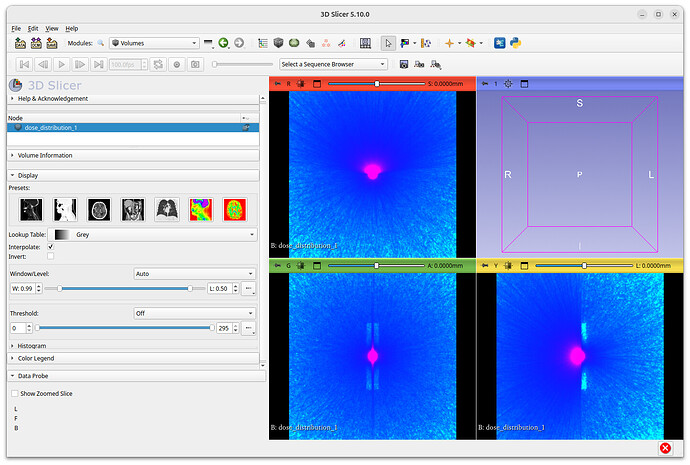
I have uploaded how the same file loads into both versions of slicer. Again, in 5.8.1 i load in as a sequence (the format I want) and in 5.10.0 it will not load as sequence so I load as volume. You can see that even though grey scale is selected, it is showing colour in 5.10.0 and when I selected other colours nothing changes.
These are the errors generated in 5.10 when this data is loaded as a sequence:
Switch to module: "Data"
Could not find list kind axis in image file
Algorithm vtkITKImageSequenceReader (0x7ff026ffe020) returned failure for request: vtkInformation (0x600003a46400)
Debug: Off
Modified Time: 379967
Reference Count: 1
Registered Events: (none)
Request: REQUEST_DATA
FROM_OUTPUT_PORT: 0
ALGORITHM_AFTER_FORWARD: 1
FORWARD_DIRECTION: 0
vtkMRMLVolumeSequenceStorageNode::ReadDataInternal: Error reading file.
vtkMRMLStorageNode::ReadData: Failed to read node dose_distribution (vtkMRMLSequenceNode1) from filename='/Users/amaga/Downloads/dose_distribution.nrrd'
AddSequence: error reading /Users/amaga/Downloads/dose_distribution.nrrd
static void qSlicerIOManager::showLoadNodesResultDialog(bool, vtkMRMLMessageCollection *) Errors occurred while loading nodes: "- Error: Loading /Users/amaga/Downloads/dose_distribution.nrrd - Error reading file.\n- Error: Loading /Users/amaga/Downloads/dose_distribution.nrrd - Failed to read node dose_distribution (vtkMRMLSequenceNode1) from filename='/Users/amaga/Downloads/dose_distribution.nrrd'\n"
Thank you!
The most help would be if we could have the actual file for testing.
If that’s not possible, then maybe pasting the nrrd file header would give us some clue (since the error says list kind axis could not be found, seeing what those axes are defined as would certainly help).
@gchampa Is this a 4D file? How did you create this file? Previous versions of Slicer made random guesses about files that had more than 3 dimensions, which sometimes were correct sometimes not. Slicer-5.10 requires unambiguous definition of axes (spatial, vector component, time, etc), mostly by relying on proper setting of the kinds field in NRRD files.
If you copy here the NRRD image header then we can give you more specific advice.
This is the header from the file provided:
NRRD0004
# Complete NRRD file format specification at:
# http://teem.sourceforge.net/nrrd/format.html
type: double
dimension: 4
space: left-posterior-superior
sizes: 2 201 201 201
space directions: none (1,0,0) (0,1,0) (0,0,1)
centerings: ??? cell cell cell
kinds: 2-vector space space space
labels: "" "x" "y" "z"
endian: little
encoding: gzip
space origin: (-100,-100,-100)
Executable:=RapidBrachyMC
Writer:=Teem
distance_unit:=mm
dose_unit:=Gy
I see the issue now. The image was attempted to be loaded as a 3D+t sequence, while it is a 3D+component volume. Since Slicer supports loading/saving of 5D data, the axis kinds checks had to be made more reliable (so that for example we can distinguish a 3D+component+time image from a 3D+time+component image).
The fix is simple: specify the axis kinds to match the actual data content. In this case, use list instead of 2-vector. You can save a numpy array into a nrrd file like this:
import numpy as np
import nrrd
img = np.zeros([201,201,201, 2])
header = {
'space': 'left-posterior-superior',
'kinds': ['domain', 'domain', 'domain', 'list'],
'labels': ["", "", "", "time"],
'units': ["", "", "", "s"],
'space origin': [-137.16099548, -36.80649948, -309.71899414],
'space directions': [[1.953125, 0., 0.], [0., 1.953125, 0.], [0., 0., 1.953125], [np.nan, np.nan, np.nan]],
'axis 3 index type': 'numeric',
'axis 3 index values': "1.2 1.4"
}
nrrd.write("c:/tmp/test.seq.nhdr", img, header)
The resulting file header will look like this:
NRRD0004
# Complete NRRD file format specification at:
# http://teem.sourceforge.net/nrrd/format.html
type: double
dimension: 4
space: left-posterior-superior
sizes: 201 201 201 2
space directions: (-1.9531249999999998,0,0) (0,-1.9531249999999998,0) (0,0,1.9531249999999998) none
kinds: domain domain domain list
labels: "" "" "" "time"
units: "" "" "" "s"
endian: little
encoding: gzip
space origin: (137.16099548000003,36.806499479999999,-309.71899414000001)
data file: test.seq.raw.gz
DataNodeClassName:=vtkMRMLScalarVolumeNode
axis 3 index type:=numeric
axis 3 index values:=1.2 1.4
For more information, check out full specification of the sequence file format and examples in the script repository.