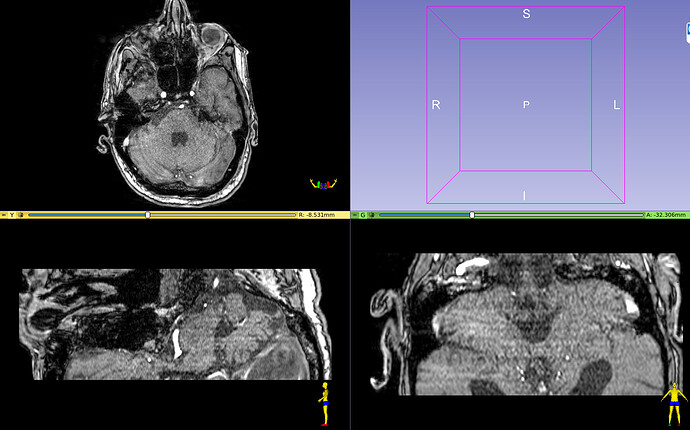
Hi All!
I have a dataset of MR angiography patients. Through sitk, I checked with GetDirections() the direction of every volume.
It turns out they all have direction (1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0), except one who has instead (1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, -1.0).
So I investigated this outlier sub with Slicer and I noticed a potential bug in the Orientation (image below).
I have 2 questions:
-
any idea of why only one sub has different Directions? Why could this be?
-
is it really a bug or am I missing something? The human in the coronal and sagittal planes doesn’t seem correctly oriented
In case you want to try it out, you can find the nifti file at this GDrive [link]
(https://drive.google.com/file/d/1VRDupP7VFHN1cLwFjzuU7t7ZnqSvL9PI/view?usp=sharing)
Thanks a lot in advance!