Bjr
j’ai des contours en format Dicom réalisé sur des images SPECT, maintenant je veux utiliser ces segments sur des images TEP/CT afin de calculer la concentration radioactives de chaque segment, je n’arrive pas à réaliser ça
pouvez vous m’aider svp!
Hi, You will need to install the SlicerRT extension to load the RTSTRUCT. I suspect that you could use the segment statistics module to calculate the average activity from the PET image, but have never tried that myself.
hi
The RT structure is already in place, the statistics don’t work, I have a table with the segment names as results only
I exported RT structure to labelmap, I got SUV values with pet SUV computation as result, however, the stats to predict volumes and other parameters still don’t work
thanks
Hi Assia, Unfortunately I also was not successful. It seems the segment statistics does not provide average intensity within a segment. You might need to write a python script.
Please use the English language in the forums for better understanding for the community. If you do not speak the language feel free to use Google Translate or any other similar service (as I suppose Greg did with your French message). Thank you!
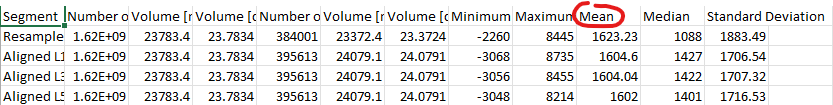
I believe it does, assuming that average and mean are the same. Here is a very recent output from the Segment Statistics module
I only get 4 columns: Segment, Surface Area, Volume (mm), Volume (cm). This is on linux. I tried 5.2.2 and 5.3.0, same result.
Interesting. It seems that you only get the statistics from the Labelmap Statistics plugin, and although all the metrics of the Scalar Volume Statistics plugin are enabled as you show in the screenshot, they are not computed. Is there any error in the console or the log? I’ll try this on Linux too when I work on that computer.
Error log completely empty when computing statistics. There were some warnings/errors on the structure import, but it seems to have loaded.
Warning: In /work/Preview/S-0-E-b/SlicerRT/DicomRtImportExport/ConversionRules/vtkPlanarContourToClosedSurfaceConversionRule.cxx, line 1258
vtkPlanarContourToClosedSurfaceConversionRule (0x116841d0): GetSpacingBetweenLines: Contour spacing is not consistent.
Warning: In /work/Preview/S-0-E-b/SlicerRT/DicomRtImportExport/ConversionRules/vtkPlanarContourToClosedSurfaceConversionRule.cxx, line 1258
vtkPlanarContourToClosedSurfaceConversionRule (0x116841d0): GetSpacingBetweenLines: Contour spacing is not consistent.
Warning: In /work/Preview/S-0-E-b/SlicerRT/DicomRtImportExport/ConversionRules/vtkPlanarContourToClosedSurfaceConversionRule.cxx, line 1258
vtkPlanarContourToClosedSurfaceConversionRule (0x116841d0): GetSpacingBetweenLines: Contour spacing is not consistent.
Warning: In /work/Preview/S-0-E-b/SlicerRT/DicomRtImportExport/ConversionRules/vtkPlanarContourToClosedSurfaceConversionRule.cxx, line 1258
vtkPlanarContourToClosedSurfaceConversionRule (0x116841d0): GetSpacingBetweenLines: Contour spacing is not consistent.
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
No input data
Yes these seem “normal”. Thanks!
I solved the problem by creating binarylabelmap in SEGMENTATION and doing an alignment between CT’s images