hello
why I always get 0 even if I use the same data?
I mean even if both data have the same characteristics
Could you share the data sets that you tested with? You can create a segmentation from one of the Slicer sample data sets if you would prefer not to share your own images/segmentations.
If you use the same data then the difference is 0, so there is no surprise there.
no, normally, when I use the same data, the dice that will be calculated must be a high value ,
normally I get a good result not 0
but I didn’t always try the same datasets, i tried several different data and I always get 0
Segment Comparison works for me in the recent versions.
Could you please share your data? The segmentation nrrd files are enough, but a Slicer scene (mrb) would be best. Thanks!
the datasets that i have are in nifti type
you can found the datasets here
https://drive.google.com/open?id=1t_gn_UvqarHlV9bFQ8EmjsaLgZh1SPtp
Thanks for the data!
For me Segment Comparison works on these segmenations. I loaded them as labelmap, then converted them to segmentation nodes, then used Segment Comparison as normal.
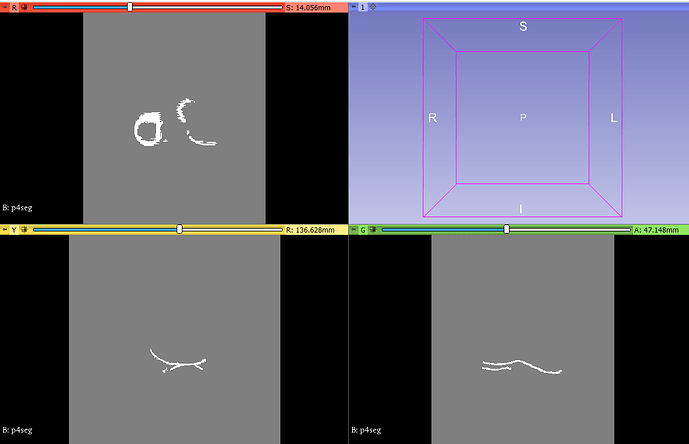
The obvious problem is that the two segmentations do not overlap, so Dice is 0 and Hausdorff distances are super high, but the algorithms work. Not sure what you did differently. Please try to do as I describe above.
can you show me how to load them as labelmap and convert them to segmentation nodes. (by a capture if you want)
because i tried to import them as labelmap but it doesn’t work
i think i made something wrong .
thak you
thank you I loaded them as labelmap, but i can’t fount the window of convertion to segmentation nodes.
As @cpinterwrote above, it is in the Data module. Make sure you use at least the latest stable version of Slicer (currently 4.10.2).
thank you it works but in Slicer 4.10.1, because in Slicer 4.10.2 i didn’t find the segment comparison module
Segment Comparison module is provided by SlicerRT extension. You’ll have the module in Slicer-4.10.2, too, if you install SlicerRT.
thank you it works
I wish I could find more function than dice and dose, such as (recall, precision, accuracy,…)
Dice, recall, precision, accuracy could be all very easily added but they are basically all useless metrics, as they are highly dependent on the shape of the segment, may be sensitive to irrelevant errors and sensitive to all kinds of clinically important errors. For example, 90% does not mean anything: a practically perfect segmentation and an extremely low quality segmentation can both get this same score. For thin shapes that I can see in your screenshots, these overlap percentage based give completely meaningless results. These metrics are probably only used because they are trivial to compute and often researchers do not want to spend a lot of time with evaluation - if they get 1-2% better values than those reported in the literature then they consider their work done, publish their paper, and move on.
Hausdorff metric variants have absolute, clinically relevant meaning - I would highly recommend to use them. Identifying anatomical landmark points and surfaces manually and measure distance between them manually gives useful information, too. You also need to spend time with carefully visual assessment of the results to see where errors occur and see if they are clinically important or not.