You can copy-paste this into the Python console to jump slice views to (0,0,0) position on Ctrl+e:
shortcut = qt.QShortcut(qt.QKeySequence('Ctrl+e'), slicer.util.mainWindow())
shortcut.connect('activated()',
lambda: slicer.modules.markups.logic().JumpSlicesToLocation(0,0,0, True))
If you want to make this keyboard shortcut permanent then copy-paste the code into your startup script
(you can find its location in menu: Edit / Application settings / General / Application startup script).
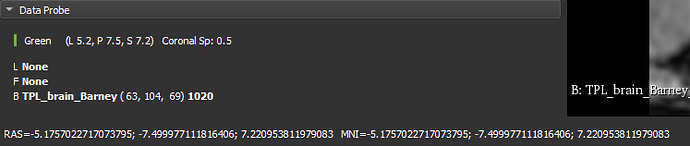
The Data probe only shows coordinate values in the world coordinate system. You can make the world coordinate system mean anything you want (e.g., MNI) by applying a transform to the volume that transforms it into that space.
You only need a small custom script (or if you want to be fancy then a custom module) to display coordinate values in multiple coordinate systems at the same time. For example, if you have a transform that moves your image to MNI space then you can use this code snippet to display world and MNI coordinates in the status bar as you move across the viewers:
def onMouseMoved(observer,eventid):
mniToWorldTransformNode = getNode('LinearTransform_3') # replace this by the name of your actual transform
worldToMniTransform = vtk.vtkGeneralTransform()
mniToWorldTransformNode.GetTransformToWorld(worldToMniTransform)
ras=[0,0,0]
mni=[0,0,0]
crosshairNode.GetCursorPositionRAS(ras)
worldToMniTransform.TransformPoint(ras, mni)
slicer.util.showStatusMessage(f"RAS={.3:ras} MNI={mni}")
crosshairNode=slicer.util.getNode('Crosshair')
observationId = crosshairNode.AddObserver(slicer.vtkMRMLCrosshairNode.CursorPositionModifiedEvent, onMouseMoved)
# Run this to stop displaying values:
# crosshairNode.RemoveObserver(observationId)
Yes, by applying a transform. You can create a transform in Transforms module and set the coordinates in Translation section, click “Invert”, and then apply this transform to the volume.
I’m not sure if it is relevant for you, but you can compute ACPC transform using ACPC transform module.