I have two 3d skull models of the same patient. I want to align them perfectly and create a model that would be their difference (skull defect).
Is this possible?
Please do not post screen capture with PHI on them.
As for your question, you can use the logical operation in the segment editor.
I edited the images. Thank you for noting it. As for the subtraction, the models are created from different ct scans and they are not aligned. I have reached this point. Is there any way to align them?
You can use the Fiducial Registration in SlicerIGT extension and use landmark sets across these to models to align them better than just using manual transforms.
I think there is a segment registration module that is built on Elastix that may help you. BUt I have no experience with that.
Manual landmark-based registration that @muratmaga recommended is a good approach that is guaranteed to work well for every case, and it as accurate as your point placement is.
If that does not work well for you for some reason then Slicer has many other registration methods. To narrow down the problem a bit we would need to know more about your constraints and requirements. How much time do you have for registration - few seconds, 1-2 minutes, 1-2 hours, or no specific time limit? How many image pairs you need to register - a few, dozens, or hundreds? How much error is tolerable - 0.1mm, about 1mm, or up to few mm?
Let me explain to you want I want to do. I have a pair of CT images for every patient (around 100 patients, a number which may be doubled). I have one CT scan before the operation and one after the procedure. The CT scans are not aligned. The surgical operation is called decompressive craniectomy and consists of removing a large piece o f bone out of the skull. What I want to do is to calculate the area of this bone defect. This area is irregular and has a curvature.
I think that there are three ways. The first one is to create a flat model without taking into account the curvature.
The second one is to create a model by comparing it with the other healthy side of the skull, like building a mirror image. (in this case, only one ct scan will be needed. I have not tried this yet)
The third is to compare the two CT scans and create the model with subtraction. This method would be the most scientific of all.
As you figured out, the problems are the alignment that affects significantly accuracy and the long time needed to create the final model.
Also, I would like to know or calculate the extend of error because of the software itself. Finally, I need a balance between time and accuracy, but I would prefer a greater accuracy method.
Is there any way for better alignment? Is it better to align the raw data from CT scans and then create the model? Maybe a script with python making tests with alignment scenarios and finding out the best could be a solution? (I have experience with vb.net; I am a “noob” in terms of python, and I don’t know how easily this could be achieved)
The model before the operation
The model after the operation
The alignment
After subtraction
The final bone piece
Based on what you describe the following will provide high enough accuracy and does not require any custom development, and can be completed in a few minutes per case (by practicing or Python-scripting the repetitive steps):
- Segment the skull on the pre-op and post-op image and export them to model nodes
- Register the pre-op and post-op CT using Fiducial registration wizard module. Up to a few mm error is acceptable (since displacement of the cut by even as much as 5-10 millimeters, would probably cause just a few percent surface area measurement error). Probably 4-6 landmarks are enough, but they need to be distributed over the head (not just on the face, but on the posterior side, too). It is worth spending some time with experimenting with which landmarks work best. Choose the pre-op image as fixed and the post-op image as moving (so that you don’t need to manually apply the transform to other nodes).
- Draw a closed contour of the craniotomy on the post-op image
- Resample the closed contour to 30-40 points, constraining control points to post-op skull surface model (Markups module / Resample). This helps preserving the contour shape when we constrain the contour to the surface in the next step.
- Constrain the closed contour to the pre-op skull surface model (Markups / Curve settings / Curve type -> Shortest distance on surface, Model node -> pre-op skull surface model).
- Cut out a curved surface patch from the pre-op skull surface model using Dynamic modeler module (add Curve cut tool, choose Model -> pre-op skull surface model, Curve -> closed curve, inside model -> new model), click Apply
- Get surface area of cut-out surface patch from Models module Information section
Once you confirmed on 5-10 patients that this workflow is good, you can automate it using Python scripting. Only the segmentation and landmark placements will need manual clicking, all the other steps can be done by a very simple, fully automatic processing script.
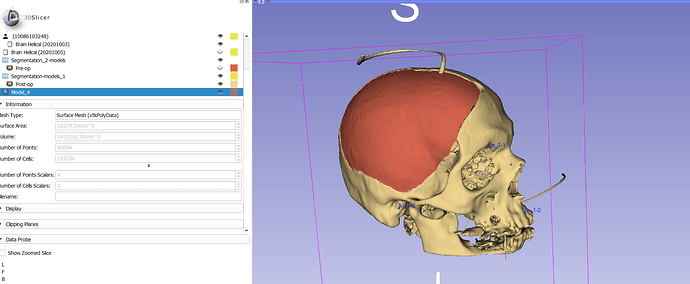
Thank you for your answer. I used your method, and I created this model (1st image) . I think that it was quite accurate. However, to have greater accuracy, firstly, I have to practice align the models with the fiducials as good as I can (3rd image), and also, when I create the curve, be as presise I can.
I also calcualted the area with the method I suggested at first. (2nd image). It was more time consuming and underestimates the area.
In case I want to use this method in a scientific article how can I asses the issue of accuracy? How can I cite the software and the relevant modules and explain the method that the algorithm uses to calculate areas?
If those are the estimates (~16,200mm2 and 15,700mm2), those differ about 3% in measurement, which doesn’t sound quite bad to me.
If you want to measure your accuracy, your best option is to do repeated measures. Meaning for every subject, you align your two meshes using landmarks, do your subtraction, and calculate the area of bone segment multiple times. From this, you can estimate the mean of the surface area for a subject and its standard error of mean. Most of this calculation can be done scripted, once you collected your sets of landmarks.
I am not sure if there is anything to cite for calculating surface area, since its simply a summation of areas of those tiny triangles. But you should cite the Slicer as a platform paper [1,2], as well as the suggested citation for SlicerIGT [3] and any other citations for the specific module(s) you are using.
[1] Fedorov, A., Beichel, R., Kalpathy-Cramer, J., Finet, J., Fillion-Robin, J.-C., Pujol, S., … Kikinis, R. (2012). 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magnetic Resonance Imaging , 30 (9), 1323–1341. doi:10.1016/j.mri.2012.05.001
[2] Kikinis, R., Pieper, S. D., & Vosburgh, K. G. (2014). 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy (pp. 277–289). Springer, New York, NY. doi:10.1007/978-1-4614-7657-3_19
[3] Ungi, T., Lasso, A., & Fichtinger, G. (2016). Open-source platforms for navigated image-guided interventions. Medical Image Analysis , 33 , 181–186. doi:10.1016/j.media.2016.06.011
I feel like there should be a way to do this at the CT volume level, but I haven’t done it–to eliminate any errors in segmenting the models from 2 different CT scans.
Another option is Meshlab–it’s not a Boolean subtraction, but you can
-
align the models with minimum 4 landmarks,
-
measure the distance between two meshes (like the Model to Model Distance module),
-
colorize the mapped model according to distance,
-
select the parts of the mesh according to distance and move them to another “layer”.
I cut a hole in a skull and ran a mock-up. I don’t know if there is a way to use the Model to Model Distance values to “select” areas the way Meshlab does.
I agree that for things that you can directly see in the image, such as diameter or circumference of the craniotomy, bone thickness, etc. you are better off working directly with the volume. However, you need segmentation if you want to get bone surface area.
There is no need for subtraction or Boolean operations, unless you need to measure bone volumes. The additional complexity with subtraction is that the two meshes are quite different (cropped differently, there are extra objects in one image that are not in the other, etc), so you would end up having to deal with many irrelevant differences.
The registration and segmentation-based workflow described above seems simple enough to me and should meet the clinical accuracy requirements.