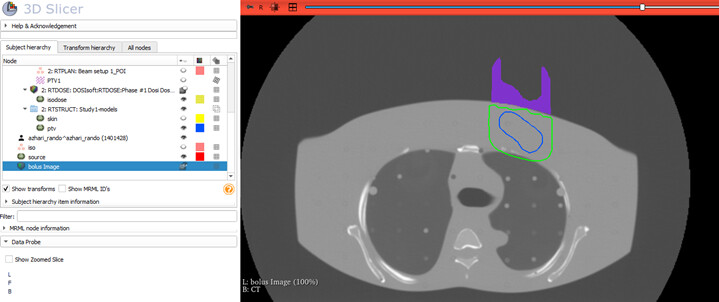
Hi professors
I manually designed individual electron bolus using 3D slicer software for my thesis a year ago.
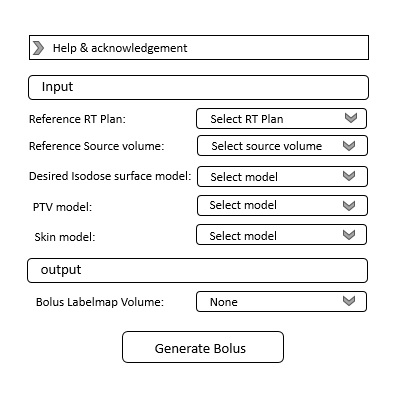
Now I have written Python codes to have electron bolus in 3D slicer automatically. and I am eager to add these codes as a module in the SlicerRT extension. I should add that note in writing these codes, I have used the SlicerSkinMouldGenerator module codes as recommended by Professor Lasso.
Unfortunately, I don’t have any experience with modules, and GUI development.
I hope developers and experts can help me convert these codes into a usable module
This module will be able to design a bolus on the top of the skin to match a desired isodose level to the distal surface of the PTV. So we can spare more normal tissue.
In the following, I will give a summary of the function of my Python scripted codes:
-
First I import the CT, RT structure (skin and ptv), RT Plan and RT dose.
-
The desired isodose model is made using the isodose module in the SlicerRT extension.
-
The RT plan metadata is accessed to obtain the isocenter and gantry angle for marking the source.
-
The points of the PTV are separated in each slice, and the source above each slice (Source_coordinate_m) is considered.
-
The farthest point of the PTV on each side (R-L coordinate) from its center is found, and a margin is applied to account for the length of bolus in each slice (mL & mR).

-
The mL and mR points are connected to Source_coordinate_m to form two lines, and then angle between two lines is found

-
The angle is divided into small angles with a step of 0.1 degree.

-
In each step, consideration is given to the line passing through the skin, PTV, and isodose.
-
putting a dot above the skin along the line equal to the distance of the intersection of the line with ptv and isodose. These dots are the border of the bolus

-
Dots are converted into poly data and then transformed into image data.
-
The image data from each slice is merged into one labelmap volume
-
Now I have a bolus with labelmap volume format
-
I can convert it to a segment or model by using 3d slicer’s capabilities.

I checked the bolus in Isogray TPS. (you can see figures in file 1 - Google Drive)
Its performance was appropriate and desired isodose surface approached the distal surface of PTV. However, in some cases at the edge of the field, it may need further handling
You can see other bolus designed for another PTV below and its performance in TPS in (file 2 - Google Drive)

.I think by these codes we can have an elementary shape of the electron bolus but it must be checked in TPS before using it
I am eager to provide you with the code I have collected and hope you can help me convert this code into a usable module.
I’m requesting help from developers and other experts to convert my codes into a usable module and integrate it into the SlicerRT extension. Additionally, I’m willing to provide the codes to anyone willing to assist me further in collaboration.
I eagerly await feedback and responses.