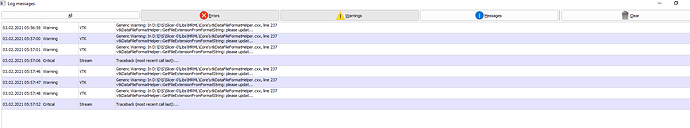
Dear Andras. Here is the message log. It’s too long. I’ve deleted some parts. Please take a look at it. Thank you very much.
“ABLTemporalBoneSegmentationModule”
[INFO][Python] 05.02.2021 19:34:26 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) - Stopping condition: Error in metric.
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Stopping condition: Error in metric.
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Stopping condition: Error in metric.
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) - Settings of AdaptiveStochasticGradientDescent in resolution 0:
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Settings of AdaptiveStochasticGradientDescent in resolution 0:
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) - Description: itk::ERROR: itk::ERROR: AdvancedMattesMutualInformationMetric(000002BF54164290): Too many samples map outside moving image buffer: 188 / 1117
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Description: itk::ERROR: itk::ERROR: AdvancedMattesMutualInformationMetric(000002BF54164290): Too many samples map outside moving image buffer: 188 / 1117
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Description: itk::ERROR: itk::ERROR: AdvancedMattesMutualInformationMetric(000002BF54164290): Too many samples map outside moving image buffer: 188 / 1117
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) - Error occurred during actual registration.
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Error occurred during actual registration.
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Error occurred during actual registration.
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) -
[INFO][Python] 05.02.2021 19:34:27 [Python] (C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py:392) - Errors occurred!
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Errors occurred!
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Errors occurred!
[INFO][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Error: Command ‘elastix’ returned non-zero exit status 1.
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - Traceback (most recent call last):
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - File “C:/Users/USER/AppData/Roaming/NA-MIC/Extensions-29402/ABLTemporalBoneSegmentation/lib/Slicer-4.11/qt-scripted-modules/ABLTemporalBoneSegmentationModule.py”, line 635, in process_transform
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - output = function()
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - File “C:/Users/USER/AppData/Roaming/NA-MIC/Extensions-29402/ABLTemporalBoneSegmentation/lib/Slicer-4.11/qt-scripted-modules/ABLTemporalBoneSegmentationModule.py”, line 812, in transform
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - log_callback=self.update_rigid_progress)
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - File “C:/Users/USER/AppData/Roaming/NA-MIC/Extensions-29402/ABLTemporalBoneSegmentation/lib/Slicer-4.11/qt-scripted-modules/ABLTemporalBoneSegmentationModule.py”, line 1318, in apply_elastix_rigid_registration
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - movingVolumeMaskNode=mask_node
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - File “C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py”, line 807, in registerVolumes
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - self.logProcessOutput(ep)
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - File “C:\Users\USER\AppData\Roaming\NA-MIC\Extensions-29402\SlicerElastix\lib\Slicer-4.11\qt-scripted-modules\Elastix.py”, line 728, in logProcessOutput
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - raise subprocess.CalledProcessError(return_code, “elastix”)
[CRITICAL][Stream] 05.02.2021 19:34:27 [] (unknown:0) - subprocess.CalledProcessError: Command ‘elastix’ returned non-zero exit status 1.