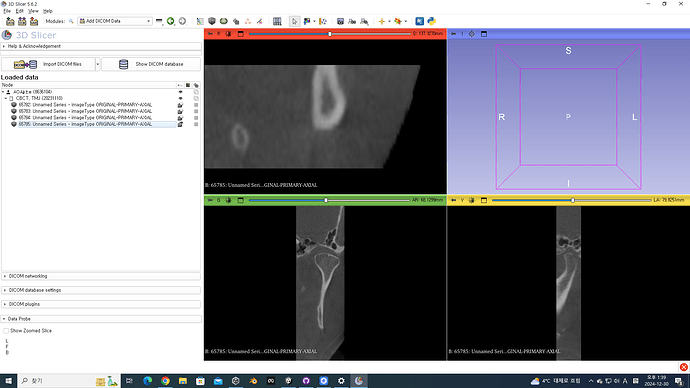
Hi,
I am trying to load a DICOM dataset into 3D Slicer, but I am encountering the following error message
This dataset works perfectly in other software like InVesalius and Mimics without any issues, but in 3D Slicer, some of the slices seem to be missing or not properly aligned after loading.
Here is some additional information about the dataset and my setup:
- The dataset is CBCT (Cone-Beam CT) data.
- I am using 3D Slicer version 5.6.2.
- In the DICOM module, the series loads as “Unnamed Series [Scalar Volume],” and the error message appears in the warnings section.
- The “Acquisition geometry regularization” is set to the default (“apply regularization transform”).
I would like to know:
- Why is this issue occurring in 3D Slicer, while the same dataset works in other software?
- Is there a specific setting or preprocessing step I should follow to fix this?
- How can I load this dataset correctly without losing any slices or data?
I have attached screenshots of the error message and the loaded volume for reference.
Any help would be greatly appreciated!
Thank you in advance!