Intro:
I need to crop teeth instances in the CBCT scan. I have to do it in Python with numpy because I need to be able to crop data during code runs and experiments - e.g., by changing the crop margin along the z-axis) - in short, it has to be done outside the Slicer. To crop instances with slicing that are not aligned with array axes, I need to rotate them first. Inverse rotation does not work as I expect it to; something is missing I need to understand.
Current process:
- In Slicer: I generate oriented bounding boxes (OBBs) based on segmentation using Slicer (I created a simple extension for it that uses class SegmentStatistics). In the end, I have an LPS JSON file per each segment with parameters like center, size, and orientation.
- Outside Slicer: I read nifti files to numpy (nibabel). I generate a matrix based on nifti affine to transform from LPS to IJK. I calculate inverse transform and apply it to bounding box vertices and CBCT scan.
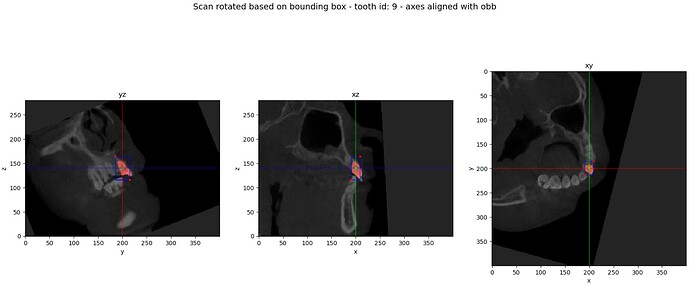
PROBLEM: I expect to obtain scan oriented in the same way as the oriented bounding box - so axes of instance are now new scan axes, so I can easily crop it using array slicing. However, I cannot get these results. Inverse rotation does not generate proper results.
Details:
I read image data from nifti file and OBBs data from JSON. The JSON file is saved in LPS, but I transform it to RAS like in here. I calculate inverse transform based on the affine matrix embedded in the nii.gz file to convert data from RAS2IJK (by IJK, I mean data space in pixels/voxels). Finally, I calculate the inverse transform of rotation in IJK. Now I can align the scan with OBB. I translate the instance center to the origin (precisely, it is in the rotation center at the beginning because of dimensions normalization and grid resampling, but I do not want to overcomplicate here), apply inverse rotation, and translate back with vector from box center to the scan center. I do the same with OBB vertices. Based on transformed vertices, I calculate slices to crop scan.
Resampling details:
I apply an affine matrix using Pytorch and MONAI. Grid resampling requires additional operation of normalizing dimensions and aligning image corners - doc: here.
Experiments and magic:
To make the transformation work, I do a bizarre and unintuitive thing. I calculate intrinsic Euler angles (‘xyz’). I change the sign of rotation around the x and y-axis. I multiply rotation matrices based on angles in proper order zyx. The magic rotation matrix constructed in this way works correctly for the scan but not for OBB vertex coordinates.
Example when it works (affine with magic):
Example when it doesn’t work (np.linalg.inv(rotationMatrixRAS)):
Slicer OBBs:
I am aware that some more explanations and details may be necessary, so please ask questions and I will try to answer them to make things clear.
*Data I am working with are publicly available.
Code:
import numpy as np
import nibabel as nib
from scipy.spatial.transform import Rotation as R
from monai.transforms import Affine
from monai.networks.utils import normalize_transform
def cropOBB(self, obbCenterLPS, obbOrientationLPS, obbSize, volumeArray, labelArray, affineRAS) -> Tuple[torch.tensor, torch.tensor, np.array, int]:
#LPS
directionLPSx = obbOrientationLPS[0::3]
directionLPSy = obbOrientationLPS[1::3]
directionLPSz = obbOrientationLPS[2::3]
obbCenterLPS4 = np.append(obbCenterLPS, 1)
rotationMatrixLPS33 = np.column_stack((directionLPSx, directionLPSy, directionLPSz))
#RAS
ras2LPS44 = np.diag([-1, -1, 1, 1])
ras2LPS33 = np.diag([-1, -1, 1])
ras2IJKinv44 = np.linalg.inv(affineRAS)
#center
obbCenterRAS4 = ras2LPS44 @ obbCenterLPS4
#rotation
rotationMatrixRAS33 = ras2LPS33 @ rotationMatrixLPS33
rotationMatrixRAS44 = np.eye(4)
rotationMatrixRAS44[:3,:3] = rotationMatrixRAS33
rotationMatrixLPS44 = np.eye(4)
rotationMatrixLPS44[:3,:3] = rotationMatrixLPS33
#obb dimensions
obbTopLeftNearRASVec3 = obbCenterRAS4[:3] - 0.5 * rotationMatrixRAS33 @ obbSize
obbBottomRightFarRASVec3 = obbCenterRAS4[:3] + 0.5 * rotationMatrixRAS33 @ obbSize
#IJK - volume data coordinates (voxels)
obbsCenterIJKVec4 = ras2IJKinv44 @ obbCenterRAS4
obbTopLeftNearIJKVec4 = ras2IJKinv44 @ np.append(obbTopLeftNearRASVec3, 1)
obbBottomRightFarIJKVec4 = ras2IJKinv44 @ np.append(obbBottomRightFarRASVec3, 1)
#transforms obb coordinates
translationOBBOriginIJK44 = np.eye(4)
translationOBBOriginIJK44[:3,3] = -obbsCenterIJKVec4[:3]
translationBack2OriginOBB= np.eye(4)
translationBack2OriginOBB[:,3] = obbsCenterIJKVec4
#translations to origin - rotation center
translation_2 = np.eye(4)
scanOriginVec3 = obbsCenterIJKVec4[:3]
translation_2[:,3] = np.append(scanOriginVec3,1) # centers tooth of interest in the screen
translation_3 = np.eye(4)
translation_4 = np.eye(4)
obbOriginVec3 = obbsCenterIJKVec4[:3] - np.array(volumeArray.shape)//2
translation_3[:,3] = np.append(-scanOriginVec3,1)
translation_4[:,3] = np.append(-obbOriginVec3,1)
#affine matrices
# norm transform - data dimensions to grid resamlt dimensions [
normTransformNonZeroCentered44 = normalize_transform(volumeArray.shape, align_corners=False, zero_centered=False)[0]
#some kind of magic
angles = R.from_matrix(np.linalg.inv(rotationMatrixLPS33)).as_euler('xyz', degrees=True)
anglesAdjusted = angles * [-1,-1,1]
rotationAdjusted = rot(anglesAdjusted[0], 'x') @ rot(anglesAdjusted[1],'y') @ rot(anglesAdjusted[2],'z')
experimental_rotation = np.eye(4)
affineMatrixNormalised44 = normTransformNonZeroCentered44 @ translation_2 @ rotationAdjusted
obbCoordinatesAffineMatrix44 = translation_4 @ translation_2 @ np.linalg.inv(rotationMatrixLPS44) @ translation_3
#apply affine obb
obbTopLeftNearIJKOriginVec4 = obbCoordinatesAffineMatrix44 @ obbTopLeftNearIJKVec4
obbBottomRightFarIJKOriginVec4 = obbCoordinatesAffineMatrix44 @ obbBottomRightFarIJKVec4
obbsCenterIJKOriginVec4 = obbCoordinatesAffineMatrix44 @ obbsCenterIJKVec4
obbCropSlices = tuple(map(lambda a, b: slice(int(np.floor(min(a,b))), int(np.ceil(max(a,b)))), obbTopLeftNearIJKOriginVec4[:3], obbBottomRightFarIJKOriginVec4[:3]))
#apply affine on volume and label and resample
volumeArray4ch= np.expand_dims(volumeArray, 0)
labelArray4ch= np.expand_dims(labelArray, 0)
transformedVolumeArray, _ = Affine(affine=affineMatrixNormalised44, normalized=True, mode='bilinear', padding_mode='zeros', image_only=False)(volumeArray4ch)
transformedLabelArray, _ = Affine(affine=affineMatrixNormalised44, normalized=True, mode='nearest', padding_mode='zeros', image_only=False)(labelArray4ch)
#crop transformed arrays
croppedToothID = transformedLabelArray[0][tuple(map(lambda a: a//2, volumeArray.shape))].int().item()
croppedVolume = transformedVolumeArray[0][obbCropSlices]
croppedLabels = transformedLabelArray[0][obbCropSlices]
#visualise results
self.visualiseResults(croppedVolume, filename=f"{croppedToothID:02d}_croppedTooth")
self.visualiseResults(transformedVolumeArray[0], filename=f"{croppedToothID:02d}_rectifiedTooth", bbox=obbCropSlices)
self.visualiseSlices(volumeArray, obbTopLeftNearIJKVec4, obbBottomRightFarIJKVec4, obbsCenterIJKVec4,
obbTopLeftNearIJKOriginVec4, obbBottomRightFarIJKOriginVec4, obbsCenterIJKOriginVec4,
transformedVolumeArray[0], transformedLabelArray[0], croppedToothID)
#delete labels of neighboring tooth
croppedLabels[croppedLabels!=croppedToothID]=0
cropAffine = np.eye(4)
cropAffine[:3,:3]=affineRAS[:3,:3]
cropAffine[:,3] = obbsCenterIJKVec4
return croppedVolume, croppedLabels, cropAffine, croppedToothID