Dear support,
I used Lung CT Segmented and Analyzer extensions and I exported lung volume from segmentation and vessels from CT Analyzer in gltf format using Open Anatomy.
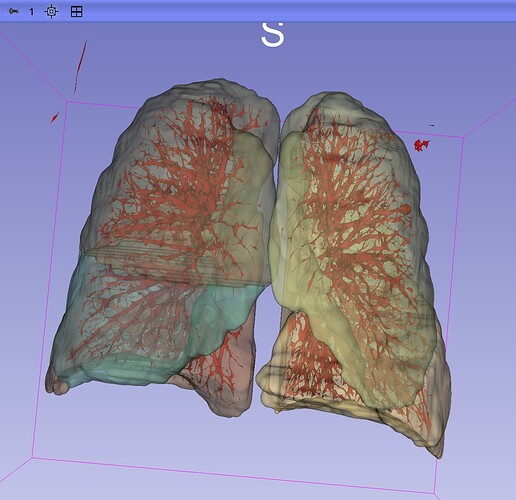
The visualization in Slicer showed good details of lung with internal vessels.
When I open the same gltf file with several online gltf viewer (https://gltf-viewer.donmccurdy.com/, glTF Sample Viewer), the models of lungs are correctly represented while the vessels models appeared as an unrealistic tetrahedral structure. what could be the problem?
Thanks
Could you please attach a few screenshots of the appearance in 3D Slicer and in the glTF viewers?
Here the screenshot of the visualization using Slicer and the gltfviewer online.
The same situation occurs when I consider lung vessels as total segmentation results.
Thanks
Thanks to the help of @lassoan, I found out glTF only supports PBR shading (Figure in the upper side).
Just other few questions:
-Why if I export only the segmentation, the lung models are correctly visible and the vessels not?
-So the correct procedure is to convert segmentation to models, set the PBR interpolation and then export in gltf format using Open Anatomy Export?
-The vessels segmentation in Slicer (using chest ct of your database) appeared with poor dimensional resolution (vessels seems to be unreal). Can I ask you the best results concerning lung vessels segmentation and what resolution and type of source dicoms should be considered?
Thank you
The gltTF file is generated the same way, regardless of the input is a segmentation or a model tree. Please compare the two files (they are just simple text files that you can open in any text editor or text comparison tool) and let us know what the differences are.
However, it would better if you used a 3D viewer that propertly supports transparent surface rendering (can do depth sorting, depth peeling, or similar methods).
What you describe works but more complex than necessary.
For high-quality vessel segmentation you need high-resolution images. Typical resolution for surgical planning is 1mm. For vessel segmentation (especially for veins) you may need contrast agent.
For your information, Slicer now offers fully automated lung, lobes, airways, and vessel segmentation. There are several active discussions about it see for example here:
Dear @lassoan,
first of all thanks for support and for informations.
I will check and share with you the differences.
What do you mean?
Thanks for sharing with me this extention. I compared the results of segmentation using: a) Lung segmentation extension and then vessels segmentation using Total segmentation and b) Monai extension. I used the same source origin (toracic volumetric with high resolution (slice thickness: 1 mm)). In both cases, if I zoom on vessels resolution, the result is not good (see picture in attach).
So, in order to have good resolution on vessels segmentation, I need contrast agent dicom?
Thanks
Hi professor, looks like I have some problem when I am exporting the segmentation to gltf, the segmetation from slicer looks like this
However, when exported to unity, the blood vessels in the gltf become like this


I tried using an online gltf viewer and it seems that there is a similar problem:
May I know what can be done to solve this problem
Thank you
For intricate structures like this, you need to reduce the decimation.
Thank you for your prompt response. I followed this video for decimation and indeed got better results.
May I also know that if there are little pieces of blood fragments near the blood vessel but they are not connected (see attached), apart from manually segmenting them out, is there a better way in the surface tool box extensions or other extensions to clean them up.
thank you very much
Small vessels are usually not needed for surgical planning and just occlude relevant parts, so commonly users apply Islands effect / Keep largest island method on the automatic vessel extraction result to get rid of them.
Thank you
Sorry for asking again. I found that for some other samples, the black spots still persist after numerous decimation, I have used larger value for decimation and tried other methods in the surface toolbox (uniform remesh, decimate, smooth, compute surface normals) If i use keep largest or island, it will only get a sub part of the vessels since they are not connected. I am still not sure why this problem happened, do you have other suggestions to solve it ?
Moreover, may I know is it possible to write a simple python script that can eliminate small fragments that are smaller than some size (e.g. 10 mm)
Thank you very much for your response.
You can use many tools in Segment Editor to get rid of small vessels but preserve large ones. It is an independent question from glTF export and is discussed in separate topics on this forum. Please read recent topics related to lung and if you have trouble producing good lung vessel segmentation then you can ask for more help by commenting in those topics.
These are all unnecessary. What I meant is to simply decrease the “Reduction factor” in the OpenAnatomy Export module to a lower value to preserve more details. If you set the slider to 0 then the exported glTF file will contain the exact same details as you see in Slicer.
Thank you for your prompt response. I found that the exported blood vessels was not displayed correctly in gltf or obj. I have tried 0.0 decimation value. I have tried different platforms, meshlab, online viewer and unity but the result is still the same.

blood vessels in unity obj

blood vessels in unity gltf

blood vessels in meshlab obj
Steps to reproduce:
Data: LIDC-IDRI-0078
Slicer 5.6.1, Slicer 5.7.0 Preview release.
Use MONAI Auto 3D seg, lung 2.0.1 to produce lung segmentation
Export to gltf using Openanatomy export
This is the expected vessels in slicer
May I know if there is any workaround?
Thank you very much
I found that the decimation process also produced some errors in slicer but was not able to resolve them.
The errors prevented the GLTF file from being opened in Unity, causing compatibility issues. If I opened the gltf file on gltf viewer online, looks like the vessel can be shown clearly but there are still a lot of errors as shown on the left bottom corner.
Fig 1: The vessels are shown correctly on gltfviewer after decimation
errors of the gltf file after decimation
On the other hand, I used the CT chest sample from slicer and created a lung segmentation using v2.0.1 MONAI 3D Auto Seg and the blood vessels have similar problem. The attached picture didn’t undergo any decimation. The result after decimation is similar too.
I use both Slicer 5.6.1 and preview release version. For the 5.6.1 version, the problem persist after decimation. For the preview release version, the display results after decimation are acceptable but with errors, those errors prevent the file from being opened in Unity.
Please check if the openanatomy export to gltf function is working correctly
Thank you very much.




![Reduce the File Size of Your 3D Model - "Decimation" [3D Slicer Workflow]](https://img.youtube.com/vi/d-0y7BHJ0oU/maxresdefault.jpg)








