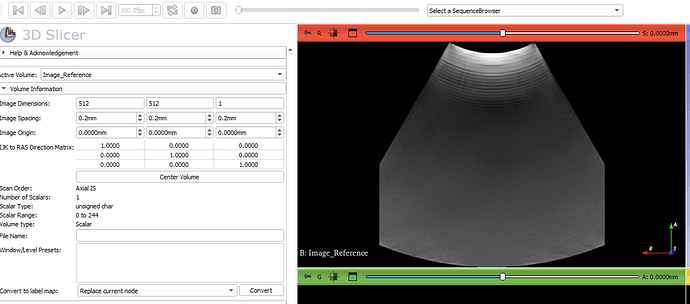
Hello I am new to 3D slicer.I have a telemed C5-2R60S-3 potable ultrasound probe and windows 11 Pro.I want to scan the abdomen and generate 3d volume.I have used plusApp & Slicer openIGTlinkIF to successfully visualize real-time ultrasound image in Slicer.When I check the details of active volume in slicer,I see pixel spacing (0.2,0.2,0.2) and image size (512,512,1).I also verified same pixel spacing(0.2,0.2,0.2) by exporting image as DICOM file and reading the DICOM metadata .
To check the correctness of pixel spacing:
From github I came to know that we can calculate image height (in mm)=total_pixels in y-axis (512)*pixel spacing in y-axis(0.2) =102.4mm.But I have set the depth of ultrasound as 150mm in echowaveII(telemed) software and I am assuming that image height should be near to 150 mm.
I need help to calculate(or find) right pixel spacing and generate multi-frame 3d reconstruction in slicer.
Any help is really appreciated.I am not able to understand how I move forward from this point i.e. without right pixel spacing.
Thank You.