I have a volume that is 15.44 um isotropic. When I loaded it into the slicer and segmented it, I did not know that the Slicer loaded it as a 1mm volume. Now I figured out the measurements are wrong. When I load the correct scale volume to Slicer and try to overlay the segment onto it, I cannot see the segments. How can I handle this without re-segmenting the whole thing again?
2D formats do not have way to store the voxel dimensions, so when you read the image sequence into Slicer, Slicer simply reports what’s in the file, often the default 1mm.
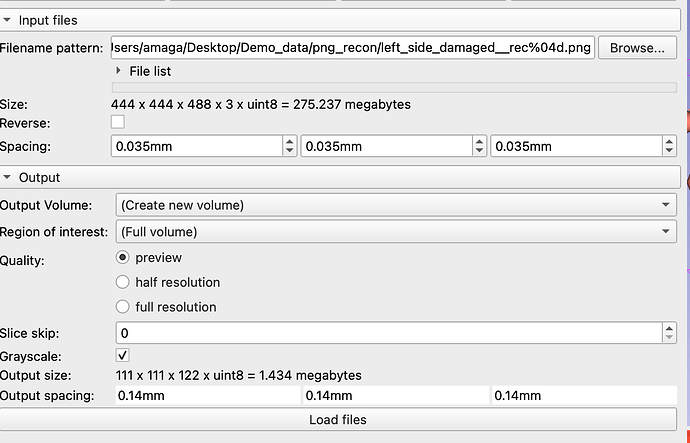
if you are working with microCT data, you should look into SlicerMorph extension, and particularly its ImageStacks module, which helps to avoid this type of issues (you can manually edit the image spacing at the time of import).
It also allows many other features such as importing sub-regions of the dataset, while preserving their spatial relationships.
If you want to correct the spacing, you should do it both for segmentaiton and the original volume. Original volume is easy, go to Volumes module and enter the correct spacing.
For segmentation, you need to first export it as a label map. Go to the segmentations module and find the export/import tab, and then export it as a labelmap. Then you can use the volumes module to edit the image spacing of the new labelmap, and then reimport it as a segmentation.
Setting the correct spacing at the time of import, or editing prior to segmentation will avoid issues like this.
Thank you. This worked ![]()