It has been possible to use Slicer from Jupyter notebooks but the most recent Slicer Preview Release takes this to a whole new level.
Click here to try a few example interactive notebooks now - or see a short video demonstrating the new features:
Highlights:
- Improved interactive use
You can now use IPython widgets (sliders, buttons, etc.) to control Slicer or adjust processing and visualization parameters. For example, CLI module parameters can be adjusted using slider widgets and results are computed and displayed autoamtically in the notebook:
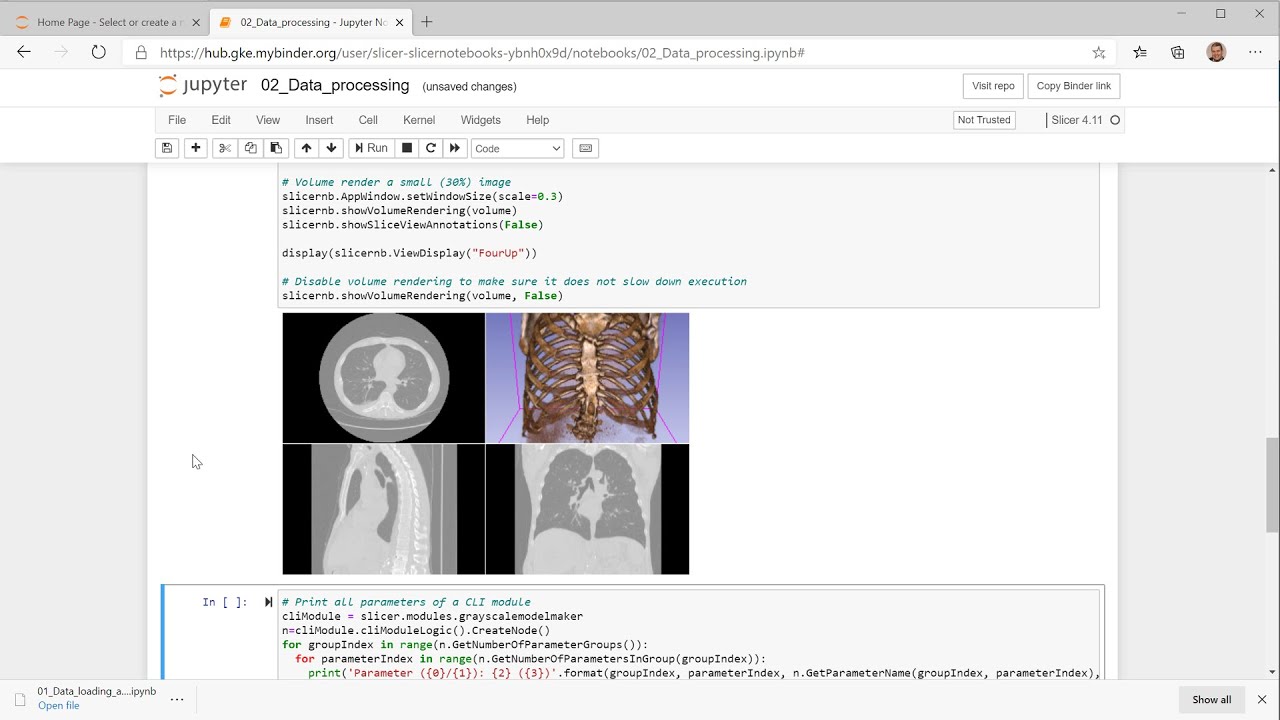
3D Slicer views are in notebooks are now interactive. You can zoom/rotate 3D views, browse slices, place points, or edit segmentations - right in the notebook:
You can display and control selected viewers, all viewers, or the entire application in notebook cells. This allows users to implement complete data processing workflows in a notebook, even if certain steps require manual user inputs (3D regions, seed points, etc.).
- Data node visualization directly in the notebook
Slicer takes advantage of rich data visualization capabilities of Jupyter notebooks. For example, markup fiducial list node is displayed as a nicely formatted table, model node is rendered as a 3D object, etc. New visualizers can be added for all MRML node types, if this proves to be useful.
- Available on the web
Since now the full application user interface is available interactively in Jupyter, it is possible to use Slicer remotely, in a web browser, without installing any software locally. For example, click here to launch Slicer in your web browser using binder - a free Jupyter web hosting service. They allocate quite limited computational resources to each instance, but it still sufficient to run Slicer.
[
Many thanks to Jupyter/xeus team, and especially to Sylvain Corlay and Martin Renou for their excellent work of creating a robust and flexible Jupyter kernel for C++ applications with embedded Python; and to @jcfr for setting up the new Slicer docker container.
More information, instructions for local installation: GitHub - Slicer/SlicerJupyter: Extension for 3D Slicer that allows the application to be used from Jupyter notebook
Any comments and suggestions are welcome.