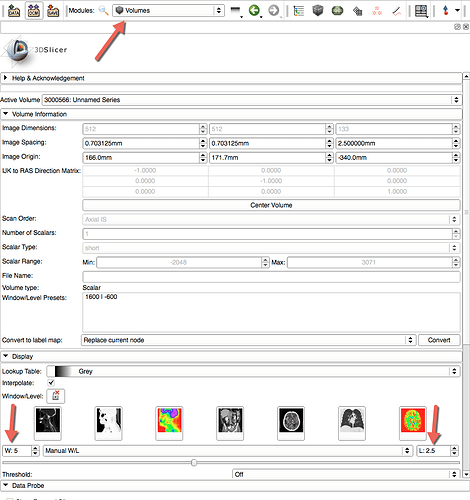
Thank you! I installed the extension and analyzed the data. This is the chart I got:
And it autogenerates a DoubleArray file with an extension .mcsv and it includes this result:
nolabels
0,5813.11,0
6.94764e+006,4719.33,0
1.38953e+007,4900.67,0
2.08429e+007,5255.11,0
2.77906e+007,5131.78,0
3.47382e+007,5241.11,0
4.16859e+007,5384.22,0
4.86335e+007,4986.33,0
5.55811e+007,5158.11,0
6.25288e+007,5173.11,0
6.94764e+007,5138.44,0
7.64241e+007,5346.67,0
8.33717e+007,6408.22,0
9.03193e+007,8021.33,0
9.7267e+007,9395.11,0
1.04215e+008,10528.4,0
1.11162e+008,11141.3,0
1.1811e+008,11415.9,0
1.25058e+008,11819.1,0
1.32005e+008,12122.1,0
1.38953e+008,12290.1,0
1.459e+008,13023.7,0
1.52848e+008,13199.8,0
1.59796e+008,13763.8,0
1.66743e+008,13793.6,0
1.73691e+008,14089.1,0
1.80639e+008,14338.1,0
1.87586e+008,15036.4,0
1.94534e+008,14694.1,0
2.01482e+008,14811.1,0
2.08429e+008,15017.1,0
2.15377e+008,15197.2,0
2.22325e+008,15255.7,0
2.29272e+008,15586.6,0
2.3622e+008,15690.2,0
2.43167e+008,15757.6,0
2.50115e+008,15632.6,0
2.57063e+008,15837.7,0
2.6401e+008,16271.6,0
2.70958e+008,16341.7,0
2.77906e+008,16426.1,0
2.84853e+008,15834.7,0
2.91801e+008,16637.7,0
2.98749e+008,16571.2,0
3.05696e+008,16795.2,0
3.12644e+008,16796.2,0
3.19592e+008,17083.7,0
3.26539e+008,17202.4,0
3.33487e+008,17062.8,0
3.40434e+008,17013.9,0
3.47382e+008,17579.1,0
3.5433e+008,17424.3,0
3.61277e+008,17485.4,0
3.68225e+008,17526.3,0
3.75173e+008,17495,0
3.8212e+008,17643.1,0
3.89068e+008,17620,0
3.96016e+008,17980,0
4.02963e+008,17697,0
4.09911e+008,17534.4,0
4.16859e+008,17951.7,0
4.23806e+008,18114.4,0
4.30754e+008,17758.4,0
4.37701e+008,17475.2,0
4.44649e+008,18047.6,0
4.51597e+008,18049.3,0
4.58544e+008,18114.2,0
4.65492e+008,18143.8,0
4.7244e+008,17907.7,0
4.79387e+008,18021.4,0
4.86335e+008,18113.4,0
4.93283e+008,18072.1,0
5.0023e+008,18285.4,0
5.07178e+008,18129,0
5.14126e+008,18383.8,0
5.21073e+008,18435.2,0
5.28021e+008,18395.6,0
5.34968e+008,18353.1,0
5.41916e+008,18423.4,0
5.48864e+008,18134.6,0
5.55811e+008,18456.9,0
5.62759e+008,18157.1,0
5.69707e+008,18588,0
5.76654e+008,18521.9,0
5.83602e+008,18377.3,0
5.9055e+008,18373.1,0
5.97497e+008,18445.3,0
6.04445e+008,18636.2,0
6.11393e+008,18641,0
6.1834e+008,18625.8,0
Is this what we are supposed to get? It`d be very helpful to see a module including the steps for analysis and the parameters.
Thanks.