Operating system:windows 10
Slicer version: 4.7.0
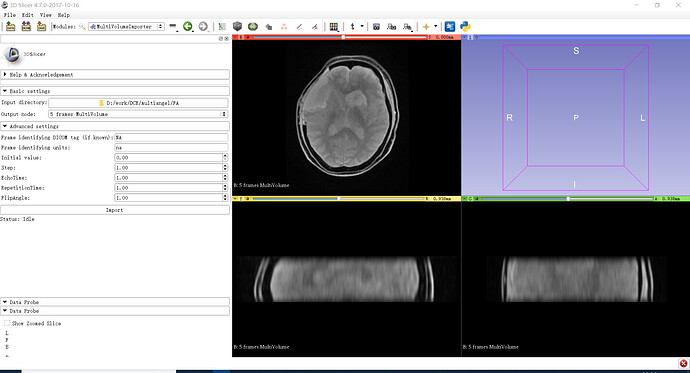
Expected behavior: output multivolume
Actual behavior: don’t know how to fill the settings, where can I get those informations? Thanks a lot!
What is the format of your input data?
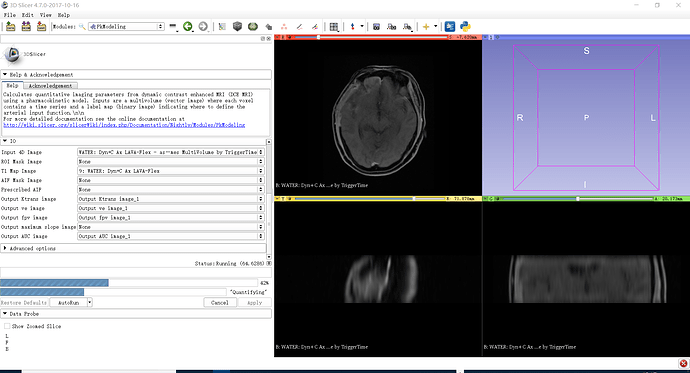
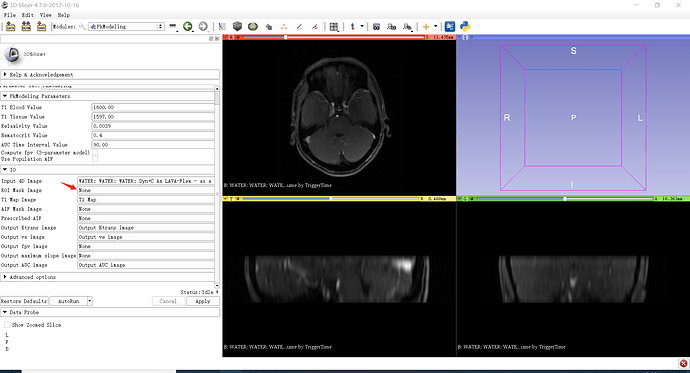
thanks a lot! I have solved the problem with somebody help, but when I use PKmodeling to calculate the Ktrans, I got the output without mask image, but when I choose the ROI mask image, there are errors. Somethings happened when I got the mask image.
Glad to hear you solved the problem. It would be nice if you could post the solution so that if someone has a similar issue your post could help them.
About your new question, please include the full error log and ideally the de-identified dataset to allow reproduce the problem.
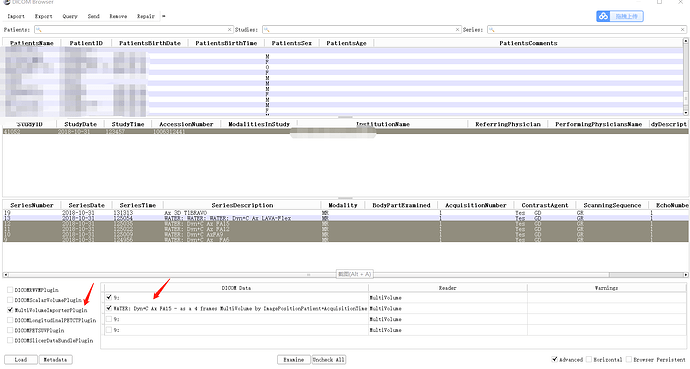
I use the Dicom module to solved the problem.
Actually I don’t know how to add the ROI image that allow me to analysis the region of interest like circle/cube/free area, and how I can get the time intensity curves?
Thanks a lot!!
Makes sense. You should use DICOM Browser to load multivolumes from DICOM. The purpose of MultiVolumeImporter is to load multivolumes that are saved as a sequence of volumes in non-DICOM format.
You first need to extract a single frame from the multivolume, and then use Editor module to segment the regions of interest. You can then plot curves averaged over those regions.
If you want to see curves for the individual pixels, you can do that using MultiVolumeExplorer module.
If this is too confusing, I will make a demo video later.
Thanks a lot, I will try it as you say.