Operating system:Windows 10
Slicer version:4.10
Expected behavior: I received a bunch of dcm files. I importem them into SLICER but I cannot see the 3D view. One one of the series, the longitudinal view has images, but the other views seem to be onlly a single line. Some other series show only saggital view as being present. the other 02 views are lines.It seems to me that the images are there but SLICER cannot get the correct Saggital, coronal and Longitudinal views in Sync. as anyone had this problem before ? HOw to solve it ?
Actual behavior:
Make sure to use the DICOM module to load your data (drag-and-drop the entire folder to the application window), import the folder, then load data from DICOM browser. See DICOM module documentation for details.
Dear Andras.
First of all let me wish you a wonderful 2019.
On the problem at hand, I have already imported the files using the DICOM module, but for whatever reason, the images refuse to appear as a 3D view.
Would you mind to take a look and let me know what is going on ?
The link below will download the folder with the images to your machine.
Thanks
Antonio
The shared folder is empty. Please check.
Hi, Andras,
Yes - You are right. My bad …
Please check new link below.
(link removed as data set might have contained patient information)
Thanks
Antonio
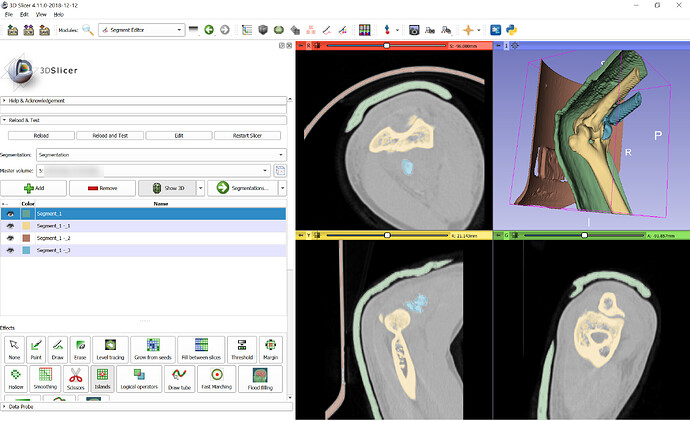
While there were some warnings logged during data loading (W: DcmItem: Length of element (7fe0,0010) is not a multiple of 2 (VR=OW)) the volume ended up loading correctly:
Note that you need to use Volume Rendering module if you want to see the image in the 3D view.