Hello! Thanks for answering
I can’t use Dcm2nii GUI but I think I identified the problem
First I converted the DICOM files to FSL and examine the bvec file and check that there is definitely a problem with the gradient table:
0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934 0.5607954357719934
-0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721 -0.5770928835459721
0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226 0.5936937619496226
All values are equal
Also in the bval file I saw that there is a problem with the b values
-4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18 -4.6116860184273879e+18
I searched in a forum for gradient tables for philips. Apparently in philips there are 3 modes: low, medium and high resolution according to the number of gradients and in turn each of them can have the “overplus” function activated or deactivated. Therefore there are six possible gradient tables. I tried several of them and got a good result with this:
0 -0.499998 -0.499998 0.707107 -0.653288 -0.208664 0.019658 0.421225 0.689925 -0.653495 -0.292882 0.294515 0.514993 0.707102 -0.707102 -0.472486 0.555492 0.707102 -0.707107 -0.707102 0.707102 0.472486 -0.707085 -0.636392 -0.706047 -0.292882 0.292882 0.707085 0.707102 -0.706260 0.034719 0.707060 0.707107 100.000.000
0 -0.499998 -0.499998 -0.707107 -0.270606 -0.675630 -0.706829 -0.567950 -0.154927 -0.270675 -0.707102 -0.706411 -0.486072 -0.292882 -0.472486 -0.707102 -0.643950 -0.472486 -0.707107 -0.472486 -0.472486 -0.707102 -0.707085 -0.425181 -0.706047 -0.707102 -0.707102 -0.707085 -0.292882 -0.706260 -0.706256 -0.707060 -0.000000 100.000.000
0 -0.707110 0.707110 0.000000 -0.707098 -0.707095 -0.707112 -0.707109 -0.707108 -0.706880 -0.643604 -0.643619 -0.706056 -0.643604 -0.526084 -0.526084 -0.526077 -0.526084 -0.000212 0.526084 0.526084 0.526084 0.007849 0.643604 0.054730 0.643604 0.643604 0.007849 0.643604 0.048932 0.707105 0.011526 0.707107 100.000.000
with that data I generated a bvec file, also generate a bval file:
0 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000 1000
Then with DWIconverter I transformed the FSL into nrrd, I created brain masking and I made the tensor estimate with good results

I made the anisotropic fraction map that showed reasonable values
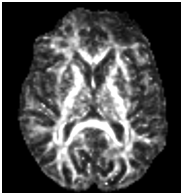
However, when I do the tractography (with the same parameters I always use) the results are not good. The tracts are aberrant.
for example in this reconstruction the roi is put in the internal capsule

something similar happens in the corpus callosum

I have read that philips gradient tables acquired with the “overplus” option sometimes require a transformation procedure but I don’t know how to do it
I’d appreciate the answer




