Hello, A few month ago I started to use 3D Slicer to create tractographys. Usually I use DWIconvert and then Difussion Brain Masking and Difussion Tensor Estimation. I’m trying to do the same but the next error comes up when I use Difussion Brain Masking: “Diffusion Brain Masking standard error:
C:/Users/Usuario/AppData/Roaming/NA-MIC/Extensions-28257/SlicerDMRI/lib/Slicer-4.10/cli-modules/DiffusionWeightedVolumeMasking.exe: Error parsing Diffusion information, no B0 images”.
After I use Difussion Tensor Estimator a black image appears.
in the volume tool the number of the scale is 17.
I hope you can give me an answer to my problem
sincerely Santiago
Hi Santiago,
As the error message says, there may be no baseline image in your data. You may want to look into that.
Also, when you look at the converted DWI data, does it look good to you?
Regards,
Fan
Hi, Thanks for answer!
The conversion from dcm to DWI its allright, I can also see the image corresponding to b0
I’ll explain the steps I followed.
This is the list of dcm files:
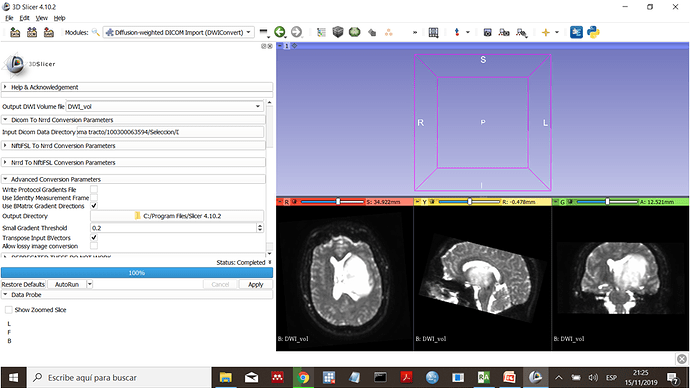
Then I use DWIconvert
The result is accurate and I can select the DWI component in the volume module
BUT… When I want to create the mask this mistake comes out:
I tried to convert dcm to nrrd with MRIcon but the same mistakes comes out again
I expect your answer!
Regards
Santiago.
Hi!
Yes, I agree that the data looks good. One thing we can check is to look at the actual b values in your data.
In conversion options, please chose DicomToFSL to output nifti format of the DWI data. In the output bval file, you will see the actual b values. Please check if there are b=0 inside.
Regards,
Fan
Hi!
I could not get the bval file with 3D slicer but I used MRIcron to create the NIfTI file and then I opened it up with DTI studio and got the next info…
I guess there is missed info in this files. Is there a way to get it back?
Yes, looks there is something wrong with the gradient table for this dataset. I am not aware if there is good way to recover. If you can another dataset scanned using the save acquisition parameters on your site. You can use the gradient table from that dataset.
Regards
Fan