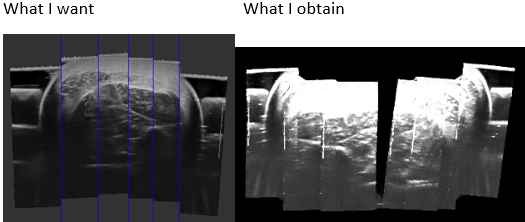
Hello,
I have 3 questions:
- Does anyone have an idea why my ultrasound images don’t match over my reconstructed volume?

I create my volume with PLUS, and the ImageToProbeTransform is included in the imageToReferenceTransform defined on the “prueba2.mha” file.
PlusApp-2.6.0.20190221-Win64\bin\VolumeReconstructor.exe --config-file=" prueba2-VolRec.xml" --image-to-reference-transform=ImageToReference --source-seq-file=" prueba2.mha" --output-volume-file=" prueba2Volume.mha"


- How I do to reconstruct my volume specifying the axes orientation:
Axes are inclined, I would like to Have the red view on the same orientation that the first ultrasound image.

Because I would like to extract SLICES from the compounding volume of ultrasound images and MASKS from the compounding volume of contours images.

If I’m not mistaken, the reconstructed volume is saved as slices throw an axis. do not?
If the reconstruction is done with the correct position and orientation of the axes, to recover the SLICES I should only read the mha format, similar to how it is done with a DICOM to obtain images. NOT?
- How I can plot the ultrasound probe like in this video:
https://www.youtube.com/watch?v=2Oc_tCu_uzs

I thanks you so much for your help
Vanessa GONZALEZ