Hi there,
This is a follow up to my previous post here
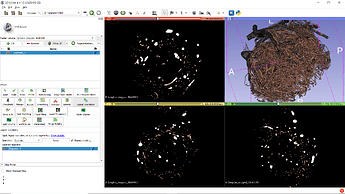
I’m trying to reconstruct a 3D volume from the 2D images provided in this study (image source).
Post reconstruction, I’d like to filter only blood vessels that range from 1-10 micrometers.
Side note: The input data for reconstructing a 3D volume is available in tiff format.
Following the instructions given in the above-mentioned link, I could obtain the following 3D volume.
 .
.
Next,
I’d like to generate a skeleton like the following (sample skeleton image below) from the 3D volume.

Next, I’d like to compute the centerline distance and maximal inscribed sphere radius (MISR) associated with each segment. By segment, I refer to the line between two branching points/ branching point and a terminal point. Previously, I have had some experience to use VMTK for obtaining centerline distance and MISR for simple skeletons like the one shown in the skeleton displayed above.
The polyData could be nicely processed further into numpy array with node numbers, edge numbers and the corresponding centerlines distances and MISR’s of each edge(/segment/branch) . So I 'd like to us the output obtained (subnetwork) from SLicer in VMTK to obtain centerline distance and MISR.
In the end, I am looking for a connected network(/skeleton), which I call as a subnetwork, with around 25 segments.
Here, I’d like to ask for suggestions on how to extract the network.
- Should I convert the 3D volume into a skeleton and then apply the constraints to extract the subnetwork.
- Or, Is it easier to extract a subvolume from the 3D volume and later skeletonize the subvolume ?
The following suggestion has been offered in the previous post
If the goal is to just extract vessels of a diameter range then it is very easy to do. If you have a segment that only contains vessels then you can use the Margin effect to shrink diameter of all vessels (this will make all vessels that have smaller diameters than the erosion disappear completely) then use the same effect to grow them back to the original diameter. This results in vessel tree that contains vessels that have diameters over the margin size (or twice the margin size - you need to check the details). To remove large-diameter vessels, you first find them using the same method, then use Logical operators effect to remove them from the image.
But I’m afraid I don’t clearly understand how to do the above after obtaining the 3D volume.
I’d like to request for help to do the above.
P.S: I’d also like to know if it’s possible to do the above programmatically. The size of the dataset is large (6GB). So I’ve been trying to work on a server by launching the GUI in X terminal. Unfortunately, it’s getting really tough, the graphics freeze and it closes when there are interruptions due to work from home:( . Doing this programmatically will allow me to use screen.